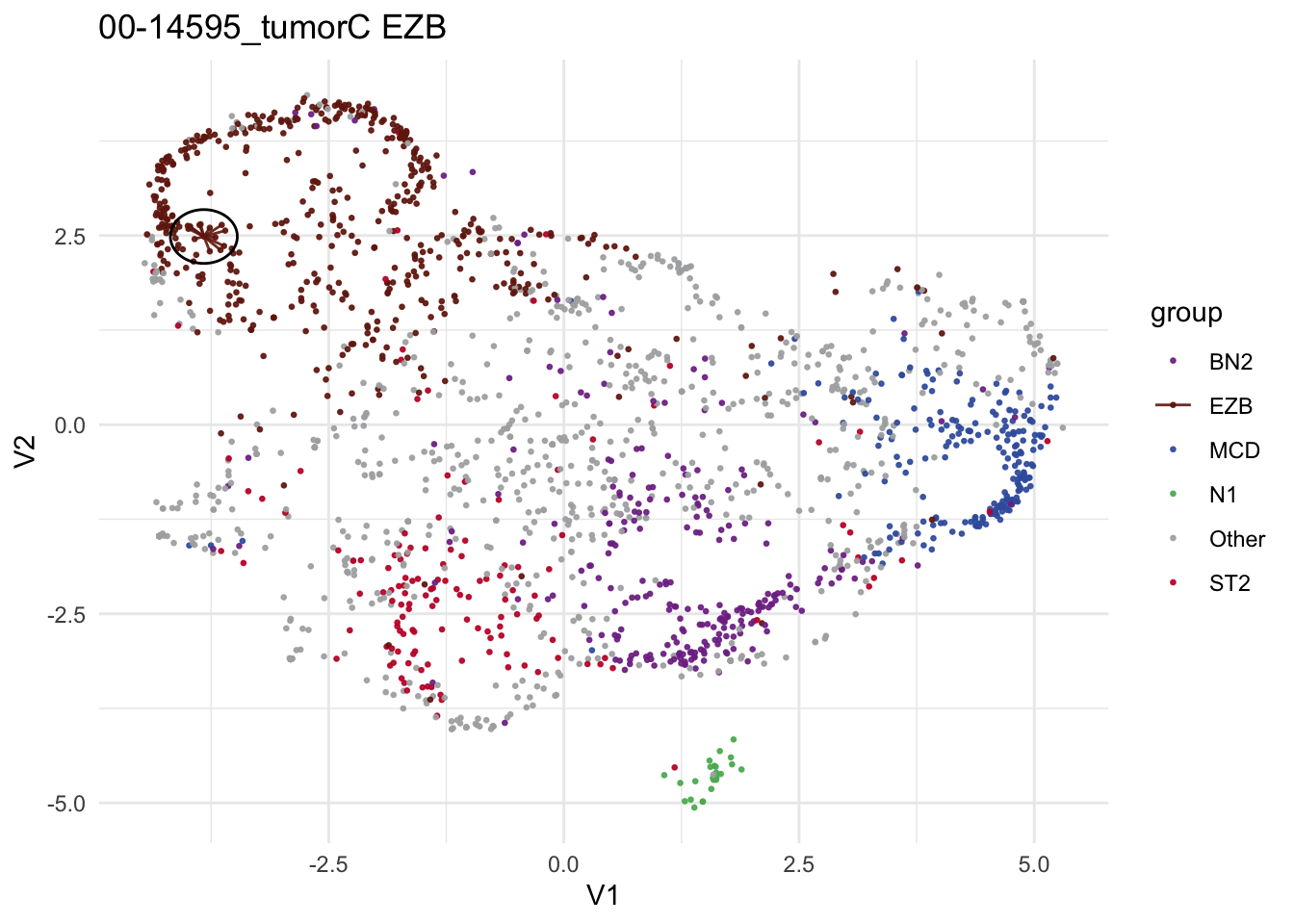
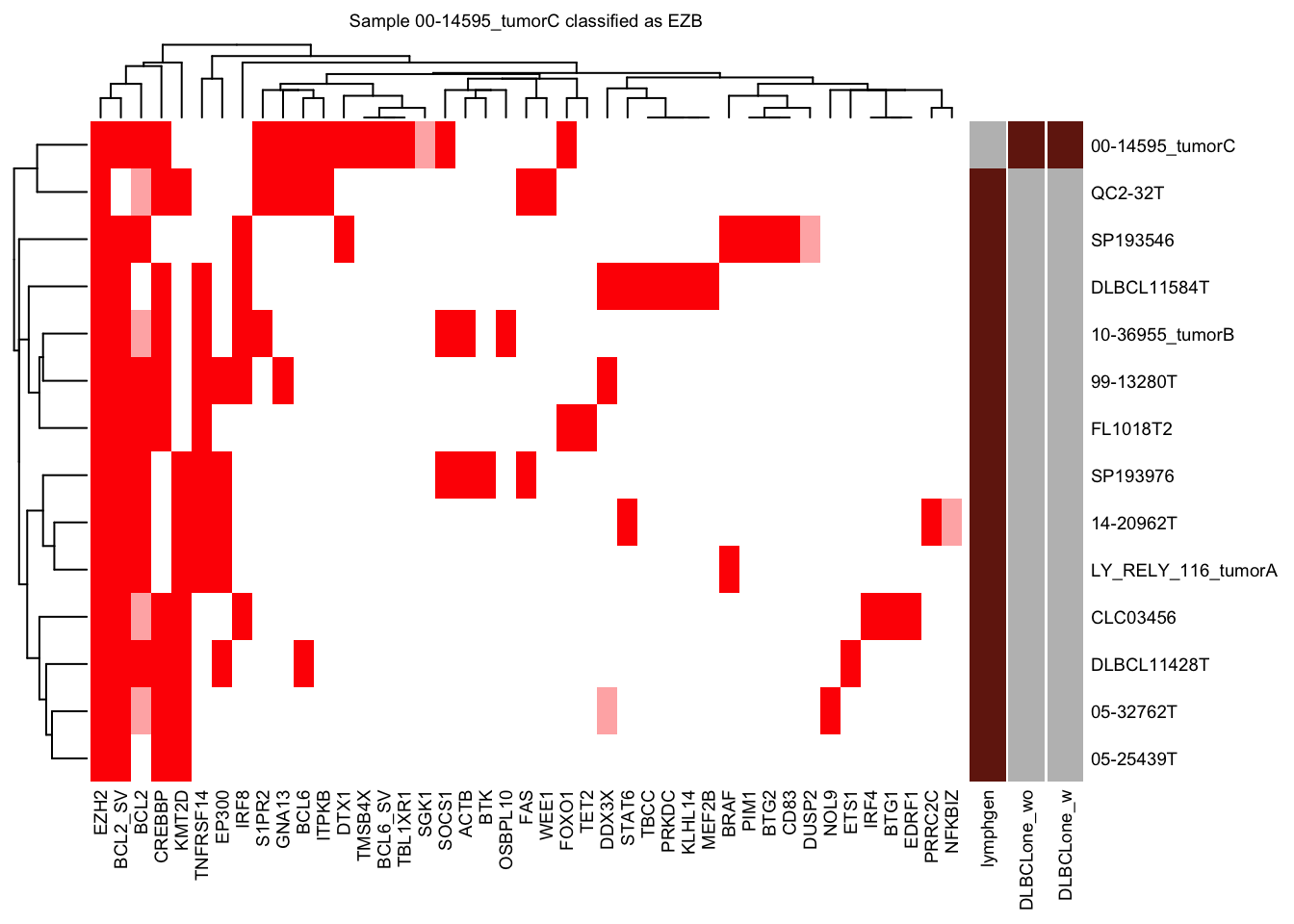
test_sample = "00-14595_tumorC"
pred_train <- DLBCLone_predict(
mutation_status = all_features_optimized$features[test_sample,],
optimized_model = all_features_optimized,
drop_extra = TRUE
)
knitr::kable(head(pred_train$prediction))| sample_id | predicted_label | confidence | other_score | neighbor_id | neighbor | distance | label | other_neighbor | vote_labels | weighted_votes | neighbors_other | neighborhood_otherness | other_weighted_votes | total_w | pred_w | V1 | V2 | .id | EZB_NN_count | MCD_NN_count | ST2_NN_count | N1_NN_count | BN2_NN_count | Other_NN_count | top_group | EZB_score | MCD_score | ST2_score | N1_score | BN2_score | Other_score | top_score_group | top_group_score | top_group_count | Other_count | by_vote | by_vote_opt | by_score | score_ratio | by_score_opt | DLBCLone_w | DLBCLone_wo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 00-14595_tumorC | EZB | 1 | 0 | DLBCL11584T,CLC03456,DLBCL11428T,05-32762T,LY_RELY_116_tumorA,99-13280T,05-25439T,QC2-32T,SP193546,SP193976,14-20962T,FL1018T2,10-36955_tumorB | 1112,351,995,85,1265,416,80,1322,1182,1164,247,89,172 | 0.203,0.223,0.231,0.259,0.265,0.293,0.297,0.306,0.321,0.34,0.345,0.349,0.352 | EZB,EZB,EZB,EZB,EZB,EZB,EZB,EZB,EZB,EZB,EZB,EZB,EZB | EZB | 46.055835675141 | 0 | 0 | 0 | 46.05584 | 46.05584 | -3.826784 | 2.487499 | 1 | 13 | 0 | 0 | 0 | 0 | 0 | EZB | 46.05584 | 0 | 0 | 0 | 0 | 0 | EZB | 46.05584 | 13 | 0 | 13 | EZB | EZB | Inf | EZB | EZB | EZB |