DLBCLone: Overview
Preface
The one classifier to classify all
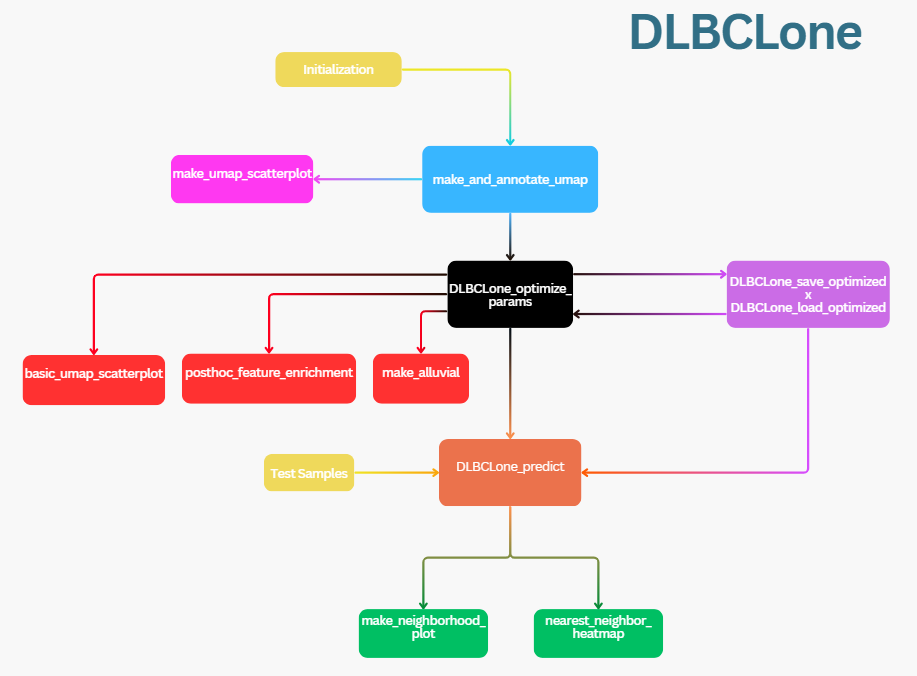
DLBCLone is a data-driven, open-source classifier for diffuse large B-cell lymphoma (DLBCL) genetic subtypes. It combines UMAP for dimensionality reduction, with a weighted k-nearest neighbors (KNN) algorithm to label samples based on neighborhood association.
The tool is designed to address key limitations of the current primary classifiers; LymphGen and DLBclass. LymphGen often labels a large proportion of samples as “Other,” while DLBclass frequently produces low-confidence predictions.
DLBCLone incorporates threshold optimization strategies to adapt to the structure of your input data. When trained on LymphGen-labeled cohorts, it can reclassify “Other” samples into genetic subtypes by leveraging their nearest neighbors in the UMAP space, providing subtype calls with associated confidence scores. See the DLBCLone preprint for more details on the methodology and its intended applications.
Introduction
This tutorial introduces how to use all of DLBCLone’s functions.
Package Requirements to run DLBCLone:
Many of the examples use the following two bundled data files.
dlbcl_meta = readr::read_tsv(system.file("extdata/dlbcl_meta_with_dlbclass.tsv",package = "GAMBLR.predict"))
dlbcl_meta_clean = filter(dlbcl_meta,
lymphgen %in% c("MCD","EZB","BN2","N1","ST2","Other"))
all_full_status = readr::read_tsv(system.file("extdata/all_full_status.tsv",package = "GAMBLR.predict")) %>%
tibble::column_to_rownames("sample_id")Alternatively, you can load the stored optimized model best_opt_model.rds from extdata:
loaded_model <- DLBCLone_load_optimized(
dirname(
system.file(
"extdata/models",
"best_opt_model.rds",
package = "GAMBLR.predict"
)
),
default_model
)
best_opt_model <- DLBCLone_activate( # Reactivates stored model setting the UMAP projection
loaded_model,
force = TRUE
)