library(GAMBLR.open)
suppressMessages(library(tidyverse))Exploring simple somatic mutations
In general, the experimental data available through GAMBLR.open is obtained using one of the get_ family of functions. These require that you specify which samples you want data from, which you accomplish by providing a metadata table that has been subset to just the samples you require. The metadata for the full set of samples available in GAMBLR.data can be obtained using get_gambl_metadata. You can subset this table using dplyr::filter. Here, we’ll focus on all DLBCL and FL samples. In many of the examples for GAMBLR.open and other packages in the GAMBLR family you will see check_and_clean_metadata. This is currently required due to the existence of near duplicate rows in the metadata. These duplicated rows exist because some samples were part of more than one study and each row refers to one of those studies. A call to check_and_clean_metadata as in this example will remove the duplicated rows, which ensures your analyses will not include duplicated data.
my_meta <- get_gambl_metadata(seq_type_filter = c("genome","capture")) %>%
dplyr::filter(
pathology %in% c("FL", "DLBCL")
)
#How many rows for each pathology and seq_type?
group_by(my_meta, seq_type, pathology) %>%
count() %>% kableExtra::kable(format="html")| seq_type | pathology | n |
|---|---|---|
| capture | DLBCL | 1783 |
| genome | DLBCL | 534 |
| genome | FL | 219 |
my_meta = check_and_clean_metadata(my_meta,duplicate_action = "keep_first")
#How many rows remain?
group_by(my_meta, seq_type, pathology) %>%
count() %>% kableExtra::kable(format="html")| seq_type | pathology | n |
|---|---|---|
| capture | DLBCL | 1783 |
| genome | DLBCL | 529 |
| genome | FL | 219 |
[1] 2531
nrow(my_meta)[1] 2531This shows that the rows in our metadata represent unique samples so we can proceed. Retrieving simple somatic mutations (SSMs) in a MAF-like format can be done a variety of ways. If your analysis is focusing on protein-coding alterations, then get_coding_ssm should meet your needs.
# retrieve MAF for all exome (capture) samples
capture_coding <- get_coding_ssm(
these_samples_metadata = my_meta,
projection = "grch37",
include_silent = TRUE,
this_seq_type = "capture"
)
nrow(capture_coding)[1] 29515
# retrieve MAF for all genome samples
genome_coding <- get_coding_ssm(
these_samples_metadata = my_meta,
projection = "grch37",
include_silent = TRUE,
this_seq_type = "genome"
)
num_genome_coding_rows = nrow(genome_coding)
genome_coding_sample = unique(genome_coding$Tumor_Sample_Barcode)
num_genome_coding_sample = length(genome_coding_sample)A total of 9875 mutations in coding regions from 546 samples were retrieved with get_coding_ssm.
To access additional mutations in non-coding regions, you can use get_ssm_by_samples if you desire all available mutations or get_ssm_by_regions if you want more control over which regions the mutations correspond to.
Note
GAMBLR.data, and thereforeGAMBLR.opendoes not contain genome-wide mutations from very many samples due to data sharing restrictions. Instead, for most samples the only non-coding mutations included are those within the regions commonly affected by aberrant somatic hypermutation (aSHM).
#retrieve genome-wide mutations for all genomes
genome_all = get_ssm_by_samples(these_samples_metadata = my_meta,
this_seq_type="genome")
num_genome_all_rows = nrow(genome_all)
genome_all_sample = unique(genome_all$Tumor_Sample_Barcode)
num_genome_all_sample = length(genome_all_sample)A total of 259732 genome-wide mutations from 594 samples were retrieved with get_ssm_by_samples.
These two approaches give us mutations from a different number of samples. We can delve into this a bit by focusing on the differences.
There are 48 sample_id that have mutations in the genome-wide result but not in coding space.
filter(my_meta, sample_id %in% genome_all_only) %>%
dplyr::select(sample_id,cohort,pairing_status, pathology, patient_id, study) %>%
kableExtra::kable(format="html")| sample_id | cohort | pairing_status | pathology | patient_id | study |
|---|---|---|---|---|---|
| 05-24561T | DLBCL_Marra | matched | DLBCL | 05-24561 | FL_Dreval |
| 14-13938T | FL_GenomeCanada | matched | FL | 14-13938 | FL_Dreval |
| 14-33798_tumorA | DLBCL_LSARP_Trios | matched | DLBCL | 14-33798 | DLBCL_Hilton |
| FL2004T1 | FL_Kridel | matched | FL | 06-25647 | FL_Dreval |
| HTMCP-01-01-00003-01D-03D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-01-00003 | DLBCL_Thomas |
| HTMCP-01-01-00012-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-01-00012 | DLBCL_Thomas |
| HTMCP-01-01-00451-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-01-00451 | DLBCL_Thomas |
| HTMCP-01-02-00013-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-02-00013 | DLBCL_Thomas |
| HTMCP-01-02-00017-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-02-00017 | DLBCL_Thomas |
| HTMCP-01-06-00036-01E | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00036 | DLBCL_Thomas |
| HTMCP-01-06-00105-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00105 | DLBCL_Thomas |
| HTMCP-01-06-00121-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00121 | DLBCL_Thomas |
| HTMCP-01-06-00136-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00136 | DLBCL_Thomas |
| HTMCP-01-06-00146-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00146 | DLBCL_Thomas |
| HTMCP-01-06-00175-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00175 | DLBCL_Thomas |
| HTMCP-01-06-00185-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00185 | DLBCL_Thomas |
| HTMCP-01-06-00206-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00206 | DLBCL_Thomas |
| HTMCP-01-06-00227-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00227 | DLBCL_Thomas |
| HTMCP-01-06-00232-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00232 | DLBCL_Thomas |
| HTMCP-01-06-00242-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00242 | DLBCL_Thomas |
| HTMCP-01-06-00253-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00253 | DLBCL_Thomas |
| HTMCP-01-06-00255-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00255 | DLBCL_Thomas |
| HTMCP-01-06-00299-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00299 | DLBCL_Thomas |
| HTMCP-01-06-00306-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00306 | DLBCL_Thomas |
| HTMCP-01-06-00307-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00307 | DLBCL_Thomas |
| HTMCP-01-06-00310-01B-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00310 | DLBCL_Thomas |
| HTMCP-01-06-00314-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00314 | DLBCL_Thomas |
| HTMCP-01-06-00419-01B-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00419 | DLBCL_Thomas |
| HTMCP-01-06-00422-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00422 | DLBCL_Thomas |
| HTMCP-01-06-00443-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00443 | DLBCL_Thomas |
| HTMCP-01-06-00485-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00485 | DLBCL_Thomas |
| HTMCP-01-06-00497-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00497 | DLBCL_Thomas |
| HTMCP-01-06-00500-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00500 | DLBCL_Thomas |
| HTMCP-01-06-00526-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00526 | DLBCL_Thomas |
| HTMCP-01-06-00563-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00563 | DLBCL_Thomas |
| HTMCP-01-06-00594-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00594 | DLBCL_Thomas |
| HTMCP-01-06-00606-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00606 | DLBCL_Thomas |
| HTMCP-01-06-00611-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00611 | DLBCL_Thomas |
| HTMCP-01-06-00634-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-06-00634 | DLBCL_Thomas |
| HTMCP-01-07-00336-01A-01E | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-07-00336 | DLBCL_Thomas |
| HTMCP-01-10-00160-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-07-00160 | DLBCL_Thomas |
| HTMCP-01-10-00778-01A-01D | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-10-00778 | DLBCL_Thomas |
| HTMCP-01-15-00366-01A-01E | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-15-00366 | DLBCL_Thomas |
| HTMCP-01-15-00367-01A-01E | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-15-00367 | DLBCL_Thomas |
| HTMCP-01-15-00370-01A-01E | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-15-00370 | DLBCL_Thomas |
| HTMCP-01-16-00265-01A-01E | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-16-00265 | DLBCL_Thomas |
| HTMCP-01-20-00272-01A-01E | DLBCL_HTMCP | matched | DLBCL | HTMCP-01-20-00272 | DLBCL_Thomas |
| SP59300 | DLBCL_ICGC | matched | DLBCL | DO27777 | FL_Dreval |
Since the non-coding mutations we get from get_ssm_by_samples are restricted to known B-cell lymphoma genes and regions affected by aSHM, we should be able to obtain most of these with a call to get_ssm_by_regions as long as the regions we request include all aSHM sites.
ashm_genome_maf = get_ssm_by_regions(these_samples_metadata = my_meta,
this_seq_type = "genome",
streamlined = F)
coding_counted = group_by(genome_coding,Tumor_Sample_Barcode) %>%
summarise(coding=n())
ashm_counted = group_by(ashm_genome_maf,Tumor_Sample_Barcode) %>%
summarise(ashm=n())
genome_all_counted = group_by(genome_all,Tumor_Sample_Barcode) %>%
summarise(all=n())
count_compare = left_join(genome_all_counted,ashm_counted)
count_compare = left_join(count_compare,coding_counted) %>%
arrange(desc(all))
count_compare = left_join(count_compare,
select(my_meta,Tumor_Sample_Barcode,cohort))
count_compare %>% kableExtra::kable(format="html")| Tumor_Sample_Barcode | all | ashm | coding | cohort |
|---|---|---|---|---|
| SU-DHL-4 | 32824 | 156 | 363 | DLBCL_cell_lines |
| OCI-Ly3 | 31532 | 120 | 353 | DLBCL_cell_lines |
| OCI-Ly10 | 30051 | 137 | 376 | DLBCL_cell_lines |
| SU-DHL-10 | 26855 | 58 | 290 | DLBCL_cell_lines |
| DOHH-2 | 22089 | 48 | 234 | DLBCL_cell_lines |
| SP192997 | 1901 | 88 | 49 | DLBCL_ICGC |
| SP116697 | 1576 | 419 | 94 | DLBCL_ICGC |
| 13-26835_tumorA | 1145 | 698 | 120 | DLBCL_LSARP_Trios |
| 01-16433_tumorB | 1051 | 92 | 36 | DLBCL_LSARP_Trios |
| SP193546 | 985 | 657 | 49 | DLBCL_ICGC |
| 16-16192T | 926 | 351 | 46 | DLBCL_GenomeCanada |
| 13-38657_tumorB | 767 | 489 | 77 | DLBCL_LSARP_Trios |
| SP193375 | 733 | 381 | 24 | DLBCL_ICGC |
| 13-38657_tumorA | 720 | 458 | 67 | DLBCL_LSARP_Trios |
| FL1019T1 | 712 | 524 | 45 | FL_Kridel |
| 10-31625T | 694 | 220 | 27 | DLBCL_GenomeCanada |
| 09-37629T | 692 | 260 | 42 | DLBCL_GenomeCanada |
| 06-11677_tumorA | 688 | 420 | 61 | DLBCL_LSARP_Trios |
| 00-14595_tumorC | 679 | 348 | 50 | DLBCL_LSARP_Trios |
| 00-14595_tumorD | 679 | 367 | 40 | DLBCL_LSARP_Trios |
| 07-35482T | 678 | 54 | 13 | DLBCL_Marra |
| 13-26835_tumorD | 668 | 331 | 67 | DLBCL_LSARP_Trios |
| SP116668 | 666 | 195 | 46 | DLBCL_ICGC |
| SP116670 | 639 | 415 | 70 | DLBCL_ICGC |
| 13-26835_tumorB | 614 | 320 | 61 | DLBCL_LSARP_Trios |
| SP124969 | 591 | 344 | 44 | DLBCL_ICGC |
| SP59304 | 582 | 208 | 39 | DLBCL_ICGC |
| SP116676 | 571 | 258 | 30 | DLBCL_ICGC |
| 17-40409_tumorB | 557 | 252 | 31 | DLBCL_LSARP_Trios |
| 17-40409_tumorA | 555 | 252 | 30 | DLBCL_LSARP_Trios |
| 16-11636T | 554 | 273 | 56 | DLBCL_GenomeCanada |
| SP59452 | 539 | 296 | 38 | DLBCL_ICGC |
| SP59448 | 525 | 237 | 30 | DLBCL_ICGC |
| 02-28397_tumorA | 524 | 296 | 41 | DLBCL_LSARP_Trios |
| HTMCP-01-06-00611-01A-01D | 510 | 512 | NA | DLBCL_HTMCP |
| SP59368 | 509 | 276 | 25 | DLBCL_ICGC |
| 14-25466T | 490 | 214 | 40 | DLBCL_GenomeCanada |
| 11-13204_tumorB | 488 | 108 | 38 | DLBCL_LSARP_Trios |
| 11-13204_tumorA | 475 | 109 | 27 | DLBCL_LSARP_Trios |
| 03-23488_tumorA | 467 | 259 | 35 | DLBCL_LSARP_Trios |
| SP116610 | 461 | 271 | 31 | DLBCL_ICGC |
| 09-12737T | 456 | 207 | 12 | DLBCL_Marra |
| SP192993 | 455 | 246 | 26 | DLBCL_ICGC |
| 03-33266_tumorB | 450 | 281 | 47 | DLBCL_LSARP_Trios |
| 07-41887_tumorA | 441 | 271 | 39 | DLBCL_LSARP_Trios |
| 16-23208T | 439 | 206 | 22 | DLBCL_GenomeCanada |
| 13-30451T | 437 | 181 | 24 | DLBCL_GenomeCanada |
| 16-18029T | 437 | 219 | 59 | DLBCL_GenomeCanada |
| FL1019T2 | 437 | 289 | 30 | FL_Kridel |
| 99-27137T | 429 | 121 | 19 | DLBCL_Marra |
| 14-35026T | 428 | 104 | 23 | DLBCL_GenomeCanada |
| 06-14634T | 417 | 169 | 12 | DLBCL_Marra |
| 11-21727T | 415 | 122 | 23 | DLBCL_Gascoyne |
| 05-15635_tumorA | 414 | 234 | 52 | DLBCL_LSARP_Trios |
| 14-41461T | 411 | 227 | 25 | DLBCL_GenomeCanada |
| 15-16885T | 404 | 184 | 20 | FL_GenomeCanada |
| SP192765 | 403 | 206 | 37 | DLBCL_ICGC |
| 13-40370T | 394 | 231 | 22 | FL_GenomeCanada |
| 15-21654T | 392 | 191 | 18 | DLBCL_GenomeCanada |
| HTMCP-01-06-00594-01A-01D | 392 | 409 | NA | DLBCL_HTMCP |
| 05-17793T | 391 | 127 | 29 | DLBCL_Gascoyne |
| 07-41887_tumorB | 391 | 228 | 35 | DLBCL_LSARP_Trios |
| FL3020T1 | 389 | 182 | 25 | FL_Kridel |
| 09-16981T | 387 | 80 | 29 | DLBCL_Gascoyne |
| FL1003T2 | 386 | 94 | 11 | FL_Kridel |
| 09-15842_tumorB | 382 | 148 | 27 | DLBCL_LSARP_Trios |
| POG707T | 382 | 173 | 16 | POG |
| 14-32442T | 378 | 215 | 32 | DLBCL_GenomeCanada |
| SP193816 | 377 | 255 | 33 | FL_ICGC |
| 07-32561_tumorB | 375 | 93 | 21 | DLBCL_LSARP_Trios |
| 01-14774_tumorA | 374 | 138 | 33 | DLBCL_LSARP_Trios |
| 09-31008_tumorA | 373 | 227 | 30 | DLBCL_LSARP_Trios |
| 07-31833T | 369 | 184 | 31 | DLBCL_Gascoyne |
| 14-27873T | 369 | 216 | 46 | DLBCL_GenomeCanada |
| 06-11677_tumorB | 365 | 238 | 22 | DLBCL_LSARP_Trios |
| 08-17645_tumorB | 364 | 196 | 35 | DLBCL_LSARP_Trios |
| LY_RELY_128_tumorA | 363 | 181 | 28 | DLBCL_LSARP_Trios |
| 09-15842_tumorA | 361 | 145 | 25 | DLBCL_LSARP_Trios |
| SP193967 | 361 | 192 | 35 | DLBCL_ICGC |
| 16-27074_tumorB | 357 | 137 | 24 | DLBCL_LSARP_Trios |
| 09-12864T | 354 | 75 | 20 | DLBCL_Gascoyne |
| 16-27074_tumorA | 353 | 135 | 23 | DLBCL_LSARP_Trios |
| SP116690 | 352 | 109 | 21 | DLBCL_ICGC |
| 16-16723T | 350 | 130 | 20 | DLBCL_GenomeCanada |
| SP193229 | 349 | 190 | 13 | FL_ICGC |
| SP192970 | 348 | 119 | 19 | DLBCL_ICGC |
| 15-43657T | 346 | 130 | 22 | DLBCL_GenomeCanada |
| 09-31601_tumorA | 342 | 150 | 21 | DLBCL_LSARP_Trios |
| 15-11617T | 338 | 110 | 28 | DLBCL_GenomeCanada |
| 15-36675T | 337 | 170 | 35 | FL_GenomeCanada |
| SP124975 | 335 | 218 | 43 | DLBCL_ICGC |
| 10-36955_tumorA | 333 | 62 | 23 | DLBCL_LSARP_Trios |
| 10-36955_tumorB | 332 | 58 | 19 | DLBCL_LSARP_Trios |
| 09-31008_tumorB | 329 | 209 | 24 | DLBCL_LSARP_Trios |
| HTMCP-01-06-00306-01A-01D | 329 | 329 | NA | DLBCL_HTMCP |
| SP193934 | 329 | 109 | 18 | DLBCL_ICGC |
| 03-23488_tumorB | 318 | 172 | 27 | DLBCL_LSARP_Trios |
| 09-33003_tumorB | 318 | 124 | 32 | DLBCL_LSARP_Trios |
| 14-37722T | 317 | 145 | 28 | DLBCL_GenomeCanada |
| SP124957 | 317 | 181 | 23 | DLBCL_ICGC |
| 04-24937T | 313 | 171 | 30 | DLBCL_GenomeCanada |
| 09-21480T | 311 | 113 | 14 | DLBCL_Gascoyne |
| 16-32248_tumorB | 311 | 114 | 22 | DLBCL_LSARP_Trios |
| 08-17645_tumorA | 309 | 186 | 28 | DLBCL_LSARP_Trios |
| 19-16466_tumorA | 306 | 165 | 14 | DLBCL_LSARP_Trios |
| FL1003T1 | 306 | 69 | 10 | FL_Kridel |
| SP124979 | 306 | 127 | 15 | FL_ICGC |
| 06-23907T | 305 | 55 | 11 | DLBCL_Marra |
| 15-41277T | 304 | 162 | 24 | FL_GenomeCanada |
| SP193025 | 302 | 137 | 15 | DLBCL_ICGC |
| SP192815 | 300 | 115 | 19 | DLBCL_ICGC |
| 09-31601_tumorB | 298 | 86 | 24 | DLBCL_LSARP_Trios |
| FL1010T2 | 297 | 118 | 27 | FL_Kridel |
| SP124971 | 294 | 101 | 19 | DLBCL_ICGC |
| SP192856 | 294 | 81 | 31 | DLBCL_ICGC |
| SP192988 | 291 | 164 | 17 | FL_ICGC |
| 03-33266_tumorA | 289 | 121 | 16 | DLBCL_LSARP_Trios |
| 05-21634T | 289 | 139 | 25 | DLBCL_Gascoyne |
| 10-39294_tumorB | 288 | 74 | 25 | DLBCL_LSARP_Trios |
| FL3011T1 | 285 | 155 | 19 | FL_Kridel |
| SP116645 | 285 | 121 | 24 | FL_ICGC |
| HTMCP-01-06-00175-01A-01D | 284 | 285 | NA | DLBCL_HTMCP |
| LY_RELY_128_tumorB | 284 | 125 | 17 | DLBCL_LSARP_Trios |
| 17-36275T | 283 | 81 | 20 | DLBCL_GenomeCanada |
| 06-19919T | 282 | 98 | 9 | DLBCL_Marra |
| FL1002T2 | 280 | 127 | 18 | FL_Kridel |
| 09-41082T | 278 | 75 | 2 | DLBCL_Marra |
| 02-15745_tumorB | 276 | 86 | 7 | DLBCL_LSARP_Trios |
| 05-18426T | 276 | 119 | 15 | DLBCL_Gascoyne |
| SP116659 | 276 | 124 | 11 | DLBCL_ICGC |
| SP192767 | 276 | 69 | 21 | DLBCL_ICGC |
| SP116657 | 274 | 61 | 19 | DLBCL_ICGC |
| 05-15635_tumorB | 272 | 124 | 15 | DLBCL_LSARP_Trios |
| FL2002T1 | 270 | 168 | 26 | FL_Kridel |
| LY_RELY_028_tumorB | 270 | 142 | 27 | DLBCL_LSARP_Trios |
| 01-14774_tumorB | 267 | 83 | 21 | DLBCL_LSARP_Trios |
| SP193420 | 267 | 191 | 11 | DLBCL_ICGC |
| 14-20552_tumorB | 266 | 147 | 27 | DLBCL_LSARP_Trios |
| 15-38154T | 266 | 69 | 15 | DLBCL_GenomeCanada |
| 15-26538T | 263 | 53 | 16 | DLBCL_GenomeCanada |
| SP116683 | 262 | 138 | 7 | FL_ICGC |
| 02-15745_tumorD | 261 | 66 | 21 | DLBCL_LSARP_Trios |
| 10-39294_tumorA | 260 | 60 | 20 | DLBCL_LSARP_Trios |
| 18-19313_tumorB | 259 | 122 | 23 | DLBCL_LSARP_Trios |
| 15-31924T | 257 | 85 | 28 | DLBCL_GenomeCanada |
| SP59400 | 256 | 77 | 16 | DLBCL_ICGC |
| 08-29440_tumorB | 255 | 127 | 28 | DLBCL_LSARP_Trios |
| 18-19313_tumorA | 254 | 120 | 23 | DLBCL_LSARP_Trios |
| 95-32141_tumorA | 254 | 84 | 22 | DLBCL_LSARP_Trios |
| SP59348 | 254 | 154 | 16 | FL_ICGC |
| 08-15460T | 253 | 128 | 40 | DLBCL_Gascoyne |
| SP116648 | 252 | 59 | 18 | DLBCL_ICGC |
| SP193766 | 252 | 112 | 16 | MALY_Other_ICGC |
| 08-15460_tumorB | 251 | 126 | 20 | DLBCL_LSARP_Trios |
| 14-35632T | 251 | 100 | 15 | DLBCL_GenomeCanada |
| 02-15745_tumorC | 248 | 71 | 20 | DLBCL_LSARP_Trios |
| SP59456 | 248 | 133 | 26 | DLBCL_ICGC |
| 17-36275_tumorB | 243 | 65 | 11 | DLBCL_LSARP_Trios |
| SP192882 | 242 | 75 | 17 | FL_ICGC |
| SP124959 | 241 | 108 | 9 | DLBCL_ICGC |
| SP192811 | 241 | 141 | 15 | FL_ICGC |
| 08-29440_tumorA | 240 | 130 | 28 | DLBCL_LSARP_Trios |
| 15-34472T | 240 | 78 | 23 | DLBCL_GenomeCanada |
| 97-18502_tumorB | 240 | 53 | 15 | DLBCL_LSARP_Trios |
| FL1018T2 | 240 | 146 | 15 | FL_Kridel |
| SP192800 | 239 | 108 | 17 | DLBCL_ICGC |
| SP194228 | 238 | 129 | 13 | DLBCL_ICGC |
| 16-43741_tumorA | 237 | 120 | 18 | DLBCL_LSARP_Trios |
| FL1002T1 | 237 | 103 | 13 | FL_Kridel |
| LY_RELY_116_tumorA | 237 | 91 | 20 | DLBCL_LSARP_Trios |
| 06-24255_tumorD | 236 | 103 | 23 | DLBCL_LSARP_Trios |
| 06-15256T | 235 | 100 | 7 | DLBCL_Marra |
| 15-30123T | 235 | 71 | 14 | FL_GenomeCanada |
| 16-27413T | 235 | 120 | 17 | DLBCL_GenomeCanada |
| SP116663 | 235 | 77 | 18 | DLBCL_ICGC |
| 10-36955_tumorD | 234 | 62 | 17 | DLBCL_LSARP_Trios |
| 14-24534_tumorB | 234 | 56 | 16 | DLBCL_LSARP_Trios |
| HTMCP-01-06-00146-01A-01D | 234 | 236 | NA | DLBCL_HTMCP |
| 09-33003T | 232 | 71 | 38 | DLBCL_Marra |
| 14-20962T | 232 | 114 | 14 | DLBCL_GenomeCanada |
| 04-38964T | 230 | 97 | 16 | FL_GenomeCanada |
| SP124973 | 229 | 118 | 18 | DLBCL_ICGC |
| 05-32947T | 227 | 20 | 8 | DLBCL_Marra |
| SP59412 | 226 | 58 | 13 | DLBCL_ICGC |
| SP193005 | 225 | 87 | 17 | DLBCL_ICGC |
| FL1020T1 | 224 | 96 | 33 | FL_Kridel |
| SP193512 | 224 | 94 | 17 | DLBCL_ICGC |
| FL1011T1 | 223 | 26 | 9 | FL_Kridel |
| SP194195 | 223 | 124 | 24 | DLBCL_ICGC |
| 02-24492_tumorA | 222 | 127 | 17 | DLBCL_LSARP_Trios |
| 07-40648_tumorA | 219 | 81 | 21 | DLBCL_LSARP_Trios |
| FL1018T1 | 218 | 140 | 14 | FL_Kridel |
| FL3003T1 | 216 | 71 | 14 | FL_Kridel |
| SP193725 | 215 | 88 | 21 | DLBCL_ICGC |
| SP194143 | 215 | 77 | 11 | DLBCL_ICGC |
| 99-13280T | 214 | 46 | 23 | DLBCL_Gascoyne |
| FL1020T2 | 214 | 94 | 31 | FL_Kridel |
| SP116649 | 214 | 14 | 8 | FL_ICGC |
| 15-18916T | 213 | 91 | 19 | FL_GenomeCanada |
| 14-33436T | 211 | 114 | 17 | DLBCL_GenomeCanada |
| HTMCP-01-06-00497-01A-01D | 211 | 211 | NA | DLBCL_HTMCP |
| 15-24306T | 210 | 91 | 18 | DLBCL_GenomeCanada |
| HTMCP-01-15-00366-01A-01E | 209 | 209 | NA | DLBCL_HTMCP |
| 00-15201_tumorA | 208 | 83 | 18 | DLBCL_LSARP_Trios |
| 07-40648_tumorB | 207 | 85 | 22 | DLBCL_LSARP_Trios |
| 05-25674T | 206 | 92 | 7 | DLBCL_Marra |
| 10-10826T | 204 | 110 | 9 | DLBCL_GenomeCanada |
| SP193976 | 203 | 96 | 29 | DLBCL_ICGC |
| SP193017 | 202 | 28 | 13 | FL_ICGC |
| 08-19764T | 201 | 51 | 10 | DLBCL_Gascoyne |
| 92-38267_tumorB | 201 | 76 | 24 | DLBCL_LSARP_Trios |
| HTMCP-01-06-00634-01A-01D | 201 | 201 | NA | DLBCL_HTMCP |
| SP116706 | 201 | 81 | 21 | FL_ICGC |
| HTMCP-01-06-00253-01A-01D | 197 | 206 | NA | DLBCL_HTMCP |
| 06-24255_tumorC | 196 | 89 | 16 | DLBCL_LSARP_Trios |
| 04-14093_tumorB | 195 | 39 | 6 | DLBCL_LSARP_Trios |
| FL1017T2 | 195 | 82 | 25 | FL_Kridel |
| HTMCP-01-06-00206-01A-01D | 194 | 195 | NA | DLBCL_HTMCP |
| 04-14093_tumorA | 193 | 37 | 6 | DLBCL_LSARP_Trios |
| 04-21856_tumorB | 193 | 95 | 19 | DLBCL_LSARP_Trios |
| 11-34915T | 193 | 128 | 14 | FL_GenomeCanada |
| 15-13383_tumorB | 193 | 88 | 13 | DLBCL_LSARP_Trios |
| 14-20552_tumorA | 192 | 100 | 22 | DLBCL_LSARP_Trios |
| 14-38639T | 192 | 123 | 22 | FL_GenomeCanada |
| 15-43891T | 192 | 88 | 15 | DLBCL_GenomeCanada |
| 95-32141_tumorB | 192 | 68 | 16 | DLBCL_LSARP_Trios |
| SP116726 | 191 | 61 | 17 | DLBCL_ICGC |
| SP192833 | 191 | 55 | 8 | DLBCL_ICGC |
| 06-30025T | 190 | 35 | 11 | DLBCL_Marra |
| 10-36955_tumorC | 190 | 58 | 13 | DLBCL_LSARP_Trios |
| 01-16433_tumorC | 189 | 82 | 14 | DLBCL_LSARP_Trios |
| 14-29443_tumorB | 189 | 47 | 11 | DLBCL_LSARP_Trios |
| FL1004T2 | 189 | 68 | 22 | FL_Kridel |
| HTMCP-01-06-00563-01A-01D | 189 | 189 | NA | DLBCL_HTMCP |
| 13-26601T | 187 | 46 | 10 | DLBCL_GenomeCanada |
| 14-29443_tumorA | 187 | 60 | 8 | DLBCL_LSARP_Trios |
| FL1004T1 | 186 | 80 | 23 | FL_Kridel |
| HTMCP-01-10-00160-01A-01D | 185 | 185 | NA | DLBCL_HTMCP |
| 16-20119T | 184 | 122 | 17 | FL_GenomeCanada |
| 11-35935T | 183 | 42 | 5 | DLBCL_GenomeCanada |
| 17-45529_tumorB | 183 | 30 | 16 | DLBCL_LSARP_Trios |
| SP59352 | 183 | 71 | 14 | FL_ICGC |
| 96-11779T | 182 | 69 | 13 | FL_GenomeCanada |
| HTMCP-01-10-00778-01A-01D | 182 | 181 | NA | DLBCL_HTMCP |
| HTMCP-01-06-00136-01A-01D | 181 | 181 | NA | DLBCL_HTMCP |
| FL3001T1 | 180 | 82 | 9 | FL_Kridel |
| 17-23504T | 179 | 71 | 12 | DLBCL_GenomeCanada |
| 15-39521T | 178 | 96 | 18 | FL_GenomeCanada |
| 02-28397_tumorB | 177 | 56 | 13 | DLBCL_LSARP_Trios |
| 04-21856_tumorA | 177 | 87 | 16 | DLBCL_LSARP_Trios |
| 14-11427T | 177 | 71 | 22 | FL_GenomeCanada |
| SP192798 | 177 | 73 | 12 | DLBCL_ICGC |
| 06-22057T | 176 | 51 | 5 | DLBCL_Marra |
| 13-34919T | 176 | 37 | 7 | FL_GenomeCanada |
| 14-23891T | 176 | 45 | 15 | DLBCL_GenomeCanada |
| SP116686 | 176 | 94 | 6 | MALY_Other_ICGC |
| SP124977 | 176 | 54 | 16 | DLBCL_ICGC |
| 15-15757T | 175 | 54 | 18 | DLBCL_GenomeCanada |
| FL1007T2 | 175 | 54 | 13 | FL_Kridel |
| HTMCP-01-06-00036-01E | 174 | 174 | NA | DLBCL_HTMCP |
| HTMCP-01-20-00272-01A-01E | 174 | 175 | NA | DLBCL_HTMCP |
| 07-25012T | 172 | 48 | 13 | DLBCL_Marra |
| SP193910 | 172 | 81 | 13 | FL_ICGC |
| 13-27960T | 171 | 115 | 19 | FL_GenomeCanada |
| 16-13732T | 170 | 40 | 6 | DLBCL_GenomeCanada |
| FL1006T2 | 169 | 78 | 14 | FL_Kridel |
| SP116701 | 169 | 50 | 9 | DLBCL_ICGC |
| SP59460 | 168 | 54 | 15 | DLBCL_ICGC |
| 04-24061_tumorB | 163 | 16 | 11 | DLBCL_LSARP_Trios |
| 89-62169T | 163 | 87 | 15 | DLBCL_Gascoyne |
| FL1016T2 | 163 | 76 | 15 | FL_Kridel |
| SP116718 | 163 | 71 | 8 | FL_ICGC |
| 07-25994_tumorB | 162 | 91 | 20 | DLBCL_LSARP_Trios |
| FL3013T1 | 162 | 80 | 4 | FL_Kridel |
| SP192850 | 161 | 39 | 14 | DLBCL_ICGC |
| 07-25994_tumorC | 160 | 89 | 21 | DLBCL_LSARP_Trios |
| 17-45529_tumorA | 160 | 26 | 13 | DLBCL_LSARP_Trios |
| SP116630 | 160 | 47 | 14 | DLBCL_ICGC |
| SP116606 | 159 | 58 | 5 | FL_ICGC |
| FL1010T1 | 158 | 58 | 8 | FL_Kridel |
| SP193258 | 158 | 80 | 16 | FL_ICGC |
| 12-32967T | 157 | 87 | 9 | FL_GenomeCanada |
| SP193543 | 157 | 48 | 13 | FL_ICGC |
| FL3009T1 | 156 | 81 | 9 | FL_Kridel |
| 15-13383T | 154 | 75 | 28 | DLBCL_GenomeCanada |
| 15-36416T | 154 | 67 | 14 | FL_GenomeCanada |
| 15-33862T | 153 | 66 | 20 | FL_GenomeCanada |
| SP192940 | 153 | 35 | 17 | DLBCL_ICGC |
| 94-15772_tumorA | 152 | 43 | 6 | DLBCL_LSARP_Trios |
| SP59312 | 152 | 42 | 10 | DLBCL_ICGC |
| 02-13135T | 150 | 49 | 11 | DLBCL_Gascoyne |
| 19-13976_tumorA | 150 | 48 | 3 | DLBCL_LSARP_Trios |
| 19-13976_tumorB | 150 | 48 | 3 | DLBCL_LSARP_Trios |
| 96-31596T | 150 | 67 | 11 | DLBCL_GenomeCanada |
| SP116674 | 150 | 76 | 14 | FL_ICGC |
| FL3008T1 | 149 | 78 | 15 | FL_Kridel |
| FL3014T1 | 148 | 58 | 9 | FL_Kridel |
| HTMCP-01-06-00242-01A-01D | 148 | 148 | NA | DLBCL_HTMCP |
| 13-26597T | 147 | 35 | 7 | FL_GenomeCanada |
| 14-34508T | 147 | 80 | 16 | FL_GenomeCanada |
| SP193326 | 147 | 48 | 14 | FL_ICGC |
| 16-10805T | 146 | 78 | 14 | FL_GenomeCanada |
| SP124963 | 146 | 93 | 14 | FL_ICGC |
| 14-11247T | 144 | 37 | 17 | DLBCL_GenomeCanada |
| 14-13959T | 144 | 33 | 8 | DLBCL_GenomeCanada |
| 16-32417T | 144 | 90 | 10 | FL_GenomeCanada |
| FL3016T1 | 144 | 79 | 12 | FL_Kridel |
| SP192870 | 144 | 65 | 19 | DLBCL_ICGC |
| 01-28152_tumorB | 143 | 37 | 14 | DLBCL_LSARP_Trios |
| HTMCP-01-15-00370-01A-01E | 143 | 144 | NA | DLBCL_HTMCP |
| SP194108 | 143 | 71 | 25 | FL_ICGC |
| 00-15201_tumorB | 142 | 44 | 15 | DLBCL_LSARP_Trios |
| 15-14583T | 142 | 50 | 11 | FL_GenomeCanada |
| 15-16852T | 142 | 45 | 14 | FL_GenomeCanada |
| 99-13520T | 142 | 44 | 20 | FL_GenomeCanada |
| 10-27154T | 141 | 39 | 12 | DLBCL_Gascoyne |
| 14-36022T | 141 | 58 | 14 | DLBCL_GenomeCanada |
| SP193364 | 141 | 57 | 5 | FL_ICGC |
| 05-22052T | 140 | 51 | 14 | DLBCL_Gascoyne |
| 05-32150_tumorB | 140 | 36 | 11 | DLBCL_LSARP_Trios |
| 15-37079T | 140 | 36 | 12 | FL_GenomeCanada |
| 14-28286T | 139 | 85 | 12 | FL_GenomeCanada |
| 14-41250T | 139 | 61 | 15 | FL_GenomeCanada |
| FL1013T2 | 139 | 29 | 6 | FL_Kridel |
| SP193040 | 137 | 57 | 7 | FL_ICGC |
| SP193914 | 137 | 36 | 11 | DLBCL_ICGC |
| 14-24907T | 136 | 54 | 11 | FL_GenomeCanada |
| 17-33596_tumorA | 136 | 23 | 3 | DLBCL_LSARP_Trios |
| SP193950 | 136 | 76 | 19 | FL_ICGC |
| FL1016T1 | 135 | 67 | 12 | FL_Kridel |
| 14-29644T | 134 | 68 | 14 | FL_GenomeCanada |
| 15-10535T | 134 | 41 | 15 | DLBCL_GenomeCanada |
| 15-15253T | 134 | 84 | 14 | FL_GenomeCanada |
| 16-29329T | 134 | 60 | 16 | DLBCL_GenomeCanada |
| SP124967 | 134 | 66 | 10 | FL_ICGC |
| FL1012T2 | 133 | 16 | 3 | FL_Kridel |
| 92-38267_tumorA | 132 | 40 | 12 | DLBCL_LSARP_Trios |
| SP116616 | 132 | 35 | 8 | FL_ICGC |
| 15-24058T | 131 | 34 | 8 | DLBCL_GenomeCanada |
| SP194077 | 131 | 56 | 16 | FL_ICGC |
| 04-24061_tumorA | 130 | 15 | 7 | DLBCL_LSARP_Trios |
| 13-19570T | 130 | 67 | 15 | FL_GenomeCanada |
| SP116618 | 130 | 50 | 11 | DLBCL_ICGC |
| SP193954 | 130 | 66 | 9 | FL_ICGC |
| 14-16707T | 129 | 38 | 3 | DLBCL_GenomeCanada |
| 17-33596_tumorB | 129 | 18 | 5 | DLBCL_LSARP_Trios |
| LY_RELY_109_tumorB | 129 | 82 | 18 | DLBCL_LSARP_Trios |
| 01-16433_tumorA | 128 | 33 | 6 | DLBCL_LSARP_Trios |
| 06-34043T | 128 | 28 | 10 | DLBCL_Marra |
| 06-30145T | 127 | 24 | 4 | DLBCL_Marra |
| 14-15505T | 127 | 64 | 12 | FL_GenomeCanada |
| 13-31210T | 126 | 56 | 11 | DLBCL_GenomeCanada |
| 15-12532T | 126 | 46 | 8 | FL_GenomeCanada |
| 03-10440_tumorB | 125 | 54 | 10 | DLBCL_LSARP_Trios |
| 14-10498_tumorB | 125 | 41 | 11 | DLBCL_LSARP_Trios |
| 14-34800T | 125 | 53 | 9 | FL_GenomeCanada |
| 17-12136T | 125 | 76 | 10 | DLBCL_GenomeCanada |
| FL1017T1 | 125 | 62 | 17 | FL_Kridel |
| HTMCP-01-16-00265-01A-01E | 125 | 125 | NA | DLBCL_HTMCP |
| 14-24648_tumorA | 124 | 30 | 5 | DLBCL_LSARP_Trios |
| 16-19402T | 124 | 52 | 10 | FL_GenomeCanada |
| SP193186 | 124 | 60 | 18 | FL_ICGC |
| HTMCP-01-06-00185-01A-01D | 123 | 123 | NA | DLBCL_HTMCP |
| HTMCP-01-06-00299-01A-01D | 123 | 123 | NA | DLBCL_HTMCP |
| 14-24648_tumorB | 122 | 31 | 6 | DLBCL_LSARP_Trios |
| FL1015T2 | 122 | 61 | 11 | FL_Kridel |
| FL3019T1 | 121 | 66 | 13 | FL_Kridel |
| 14-35472_tumorB | 120 | 23 | 9 | DLBCL_LSARP_Trios |
| 14-32185T | 119 | 48 | 14 | FL_GenomeCanada |
| FL3006T1 | 118 | 47 | 9 | FL_Kridel |
| HTMCP-01-06-00307-01A-01D | 118 | 119 | NA | DLBCL_HTMCP |
| 05-24395T | 117 | 25 | 4 | DLBCL_Marra |
| 95-32814T | 117 | 43 | 4 | DLBCL_Marra |
| 14-33262T | 115 | 39 | 13 | DLBCL_GenomeCanada |
| 01-23117_tumorB | 114 | 27 | 13 | DLBCL_LSARP_Trios |
| 14-30670T | 114 | 42 | 4 | FL_GenomeCanada |
| 16-31791T | 114 | 40 | 8 | DLBCL_GenomeCanada |
| FL3004T1 | 114 | 46 | 5 | FL_Kridel |
| 14-37865T | 113 | 41 | 7 | FL_GenomeCanada |
| SP193120 | 113 | 56 | 11 | FL_ICGC |
| SP193945 | 113 | 18 | 9 | MALY_Other_ICGC |
| 10-40676T | 112 | 44 | 6 | DLBCL_GenomeCanada |
| FL1009T1 | 112 | 42 | 10 | FL_Kridel |
| SP116635 | 112 | 53 | 8 | DLBCL_ICGC |
| 06-25674T | 111 | 33 | 4 | DLBCL_Marra |
| 07-30628T | 111 | 31 | 6 | DLBCL_Marra |
| SP116654 | 111 | 55 | 17 | FL_ICGC |
| 13-43956T | 110 | 56 | 13 | FL_GenomeCanada |
| 01-28152_tumorA | 109 | 32 | 12 | DLBCL_LSARP_Trios |
| 14-34590T | 109 | 47 | 3 | FL_GenomeCanada |
| 16-18623T | 109 | 46 | 7 | DLBCL_GenomeCanada |
| FL1006T1 | 109 | 51 | 13 | FL_Kridel |
| FL1013T1 | 109 | 28 | 5 | FL_Kridel |
| 05-24904T | 108 | 27 | 6 | DLBCL_Marra |
| SP116604 | 108 | 45 | 6 | FL_ICGC |
| FL1008T2 | 107 | 32 | 16 | FL_Kridel |
| FL3010T1 | 107 | 56 | 12 | FL_Kridel |
| HTMCP-01-06-00419-01B-01D | 107 | 107 | NA | DLBCL_HTMCP |
| SP116709 | 107 | 48 | 11 | DLBCL_ICGC |
| 98-22532T | 106 | 35 | 8 | DLBCL_Marra |
| FL1012T1 | 106 | 17 | 5 | FL_Kridel |
| SP116688 | 106 | 30 | 19 | DLBCL_ICGC |
| SP124981 | 106 | 32 | 3 | DLBCL_ICGC |
| SP193855 | 106 | 20 | 11 | FL_ICGC |
| 06-22314_tumorB | 105 | 17 | 7 | DLBCL_LSARP_Trios |
| 15-30563T | 105 | 50 | 9 | FL_GenomeCanada |
| 01-23117_tumorA | 103 | 33 | 9 | DLBCL_LSARP_Trios |
| 05-24006T | 103 | 41 | 8 | DLBCL_Marra |
| 14-32922T | 103 | 35 | 10 | FL_GenomeCanada |
| 05-32150T | 102 | 32 | 12 | DLBCL_Gascoyne |
| 15-13365T | 102 | 36 | 8 | DLBCL_GenomeCanada |
| SP59416 | 101 | 39 | 8 | FL_ICGC |
| 05-23110T | 100 | 19 | 9 | DLBCL_Marra |
| 05-12939T | 97 | 19 | 4 | DLBCL_Marra |
| 15-42543T | 97 | 54 | 7 | FL_GenomeCanada |
| 16-30371T | 97 | 48 | 7 | FL_GenomeCanada |
| HTMCP-01-06-00105-01A-01D | 97 | 98 | NA | DLBCL_HTMCP |
| 00-26427_tumorC | 96 | 24 | 14 | DLBCL_LSARP_Trios |
| 03-10440_tumorA | 96 | 52 | 10 | DLBCL_LSARP_Trios |
| 05-25439T | 96 | 12 | 4 | DLBCL_Marra |
| 06-10398T | 95 | 29 | 5 | DLBCL_Marra |
| 11-28845T | 95 | 37 | 12 | FL_GenomeCanada |
| FL3015T1 | 95 | 34 | 14 | FL_Kridel |
| SP194083 | 94 | 56 | 5 | FL_ICGC |
| 13-22818T | 92 | 26 | 9 | DLBCL_GenomeCanada |
| SP193655 | 92 | 45 | 6 | FL_ICGC |
| SP194212 | 92 | 31 | 6 | FL_ICGC |
| 07-34776T | 91 | 20 | 13 | FL_GenomeCanada |
| 14-26632T | 91 | 35 | 11 | FL_GenomeCanada |
| 15-29858T | 91 | 26 | 8 | DLBCL_GenomeCanada |
| 15-37466T | 91 | 46 | 9 | FL_GenomeCanada |
| 04-14066_tumorB | 90 | 26 | 8 | DLBCL_LSARP_Trios |
| 06-16716T | 90 | 14 | 2 | DLBCL_Marra |
| 06-22314_tumorA | 89 | 18 | 6 | DLBCL_LSARP_Trios |
| 16-27229T | 89 | 47 | 9 | FL_GenomeCanada |
| 01-20260T | 88 | 31 | 12 | FL_GenomeCanada |
| 15-14453T | 88 | 44 | 8 | FL_GenomeCanada |
| 97-18502_tumorA | 88 | 32 | 7 | DLBCL_LSARP_Trios |
| SP116638 | 88 | 41 | 6 | FL_ICGC |
| 81-52884T | 87 | 14 | 2 | DLBCL_Marra |
| SP193744 | 87 | 27 | 6 | FL_ICGC |
| FL2005T1 | 86 | 39 | 3 | FL_Kridel |
| SP194134 | 86 | 15 | 11 | FL_ICGC |
| SP59360 | 86 | 6 | 6 | DLBCL_ICGC |
| 02-20170T | 85 | 15 | 10 | DLBCL_Marra |
| SP193925 | 85 | 37 | 9 | FL_ICGC |
| 04-29264T | 84 | 39 | 3 | DLBCL_Marra |
| 13-40593T | 84 | 42 | 8 | FL_GenomeCanada |
| 14-11009T | 84 | 28 | 7 | FL_GenomeCanada |
| 15-10675T | 84 | 39 | 13 | FL_GenomeCanada |
| 15-29305T | 84 | 26 | 12 | FL_GenomeCanada |
| 15-39657T | 84 | 32 | 8 | FL_GenomeCanada |
| FL2003T1 | 84 | 9 | 4 | FL_Kridel |
| SP116723 | 84 | 39 | 12 | FL_ICGC |
| 14-11777T | 83 | 31 | 13 | FL_GenomeCanada |
| 14-32899T | 83 | 13 | 10 | FL_GenomeCanada |
| SP116720 | 83 | 26 | 7 | FL_ICGC |
| 09-11467T | 82 | 36 | 7 | DLBCL_Gascoyne |
| FL1007T1 | 82 | 33 | 8 | FL_Kridel |
| SP59340 | 82 | 34 | 9 | FL_ICGC |
| 06-24915T | 81 | 17 | 2 | DLBCL_Marra |
| 14-35472_tumorA | 81 | 17 | 9 | DLBCL_LSARP_Trios |
| SP124984 | 81 | 39 | 5 | FL_ICGC |
| SP194216 | 81 | 37 | 7 | DLBCL_ICGC |
| FL2006T1 | 80 | 35 | 10 | FL_Kridel |
| 10-39333T | 79 | 27 | 4 | FL_GenomeCanada |
| FL2008T1 | 79 | 13 | 9 | FL_Kridel |
| 04-14066_tumorA | 77 | 22 | 7 | DLBCL_LSARP_Trios |
| 94-15772_tumorB | 77 | 26 | 3 | DLBCL_LSARP_Trios |
| SP59300 | 77 | 12 | NA | DLBCL_ICGC |
| 14-13213T | 76 | 41 | 7 | FL_GenomeCanada |
| SP193808 | 75 | 14 | 6 | FL_ICGC |
| SP194043 | 75 | 32 | 8 | FL_ICGC |
| SP59356 | 75 | 27 | 6 | FL_ICGC |
| 02-22991T | 74 | 16 | 5 | DLBCL_Marra |
| 15-40296T | 74 | 35 | 7 | FL_GenomeCanada |
| 14-10498_tumorA | 72 | 27 | 5 | DLBCL_LSARP_Trios |
| SP193205 | 72 | 30 | 7 | FL_ICGC |
| SP193570 | 72 | 20 | 4 | FL_ICGC |
| SP193720 | 72 | 37 | 7 | FL_ICGC |
| 14-27524T | 71 | 29 | 5 | FL_GenomeCanada |
| SP194080 | 71 | 19 | 5 | DLBCL_ICGC |
| SP59308 | 71 | 39 | 8 | FL_ICGC |
| SP116624 | 70 | 30 | 1 | DLBCL_ICGC |
| 14-16281T | 69 | 18 | 5 | DLBCL_GenomeCanada |
| 14-35030T | 69 | 19 | 11 | FL_GenomeCanada |
| 16-37777T | 69 | 34 | 7 | FL_GenomeCanada |
| FL3007T1 | 69 | 17 | 7 | FL_Kridel |
| SP59420 | 69 | 27 | 3 | FL_ICGC |
| 15-17849T | 68 | 8 | 4 | FL_GenomeCanada |
| 16-13504T | 68 | 34 | 5 | FL_GenomeCanada |
| SP193965 | 68 | 28 | 3 | FL_ICGC |
| SP194173 | 68 | 25 | 5 | FL_ICGC |
| SP59316 | 68 | 37 | 5 | FL_ICGC |
| FL1008T1 | 67 | 27 | 14 | FL_Kridel |
| FL2001T1 | 67 | 34 | 5 | FL_Kridel |
| FL3005T1 | 67 | 36 | 7 | FL_Kridel |
| FL3017T1 | 67 | 28 | 7 | FL_Kridel |
| 06-11535T | 66 | 14 | 6 | DLBCL_Marra |
| HTMCP-01-01-00451-01A-01D | 66 | 66 | NA | DLBCL_HTMCP |
| SP194053 | 66 | 24 | 6 | DLBCL_ICGC |
| 02-15630_tumorA | 65 | 20 | 5 | DLBCL_LSARP_Trios |
| HTMCP-01-06-00314-01A-01D | 65 | 65 | NA | DLBCL_HTMCP |
| SP193347 | 65 | 5 | 6 | FL_ICGC |
| SP193650 | 65 | 31 | 9 | FL_ICGC |
| 00-26427_tumorA | 63 | 10 | 5 | DLBCL_LSARP_Trios |
| 13-29091T | 61 | 18 | 7 | FL_GenomeCanada |
| 14-33798_tumorB | 61 | 14 | 4 | DLBCL_LSARP_Trios |
| SP116627 | 61 | 17 | 7 | DLBCL_ICGC |
| SP193057 | 61 | 27 | 4 | FL_ICGC |
| SP193354 | 61 | 29 | 5 | FL_ICGC |
| SP193528 | 61 | 19 | 2 | DLBCL_ICGC |
| 06-33777T | 60 | 14 | 5 | DLBCL_Marra |
| 14-34708T | 59 | 18 | 3 | FL_GenomeCanada |
| FL3002T1 | 59 | 23 | 10 | FL_Kridel |
| HTMCP-01-06-00310-01B-01D | 59 | 59 | NA | DLBCL_HTMCP |
| SP59464 | 59 | 23 | 5 | FL_ICGC |
| 02-15630_tumorB | 58 | 16 | 2 | DLBCL_LSARP_Trios |
| HTMCP-01-06-00443-01A-01D | 58 | 58 | NA | DLBCL_HTMCP |
| SP193093 | 58 | 21 | 8 | FL_ICGC |
| SP59280 | 58 | 10 | 13 | DLBCL_ICGC |
| SP59292 | 58 | 23 | 8 | FL_ICGC |
| 09-31233T | 57 | 22 | 4 | DLBCL_GenomeCanada |
| 16-32248_tumorA | 57 | 20 | 5 | DLBCL_LSARP_Trios |
| FL1014T1 | 57 | 13 | 8 | FL_Kridel |
| FL3018T1 | 57 | 29 | 5 | FL_Kridel |
| HTMCP-01-06-00227-01A-01D | 57 | 57 | NA | DLBCL_HTMCP |
| 08-25894T | 56 | 17 | 1 | DLBCL_Marra |
| 14-13480T | 56 | 12 | 5 | FL_GenomeCanada |
| SP194238 | 56 | 25 | 10 | FL_ICGC |
| FL1001T2 | 55 | 10 | 8 | FL_Kridel |
| 14-29140T | 54 | 19 | 3 | FL_GenomeCanada |
| FL2007T1 | 54 | 22 | 7 | FL_Kridel |
| SP192804 | 54 | 13 | 6 | FL_ICGC |
| FL1015T1 | 53 | 22 | 7 | FL_Kridel |
| 16-17861T | 52 | 7 | 6 | DLBCL_GenomeCanada |
| 15-14813T | 51 | 5 | 9 | FL_GenomeCanada |
| HTMCP-01-06-00422-01A-01D | 51 | 51 | NA | DLBCL_HTMCP |
| SP59320 | 51 | 12 | 4 | FL_ICGC |
| SP59432 | 51 | 17 | 6 | FL_ICGC |
| 02-18356_tumorA | 50 | 3 | 5 | DLBCL_LSARP_Trios |
| SP193467 | 50 | 19 | 5 | FL_ICGC |
| SP193684 | 50 | 12 | 3 | DLBCL_ICGC |
| SP193801 | 50 | 23 | 4 | FL_ICGC |
| SP193828 | 50 | 15 | 7 | FL_ICGC |
| FL3012T1 | 49 | 11 | 7 | FL_Kridel |
| SP193992 | 49 | 21 | 5 | FL_ICGC |
| SP194065 | 49 | 13 | 8 | FL_ICGC |
| 02-18356_tumorB | 48 | 6 | 7 | DLBCL_LSARP_Trios |
| FL1005T2 | 48 | NA | 5 | FL_Kridel |
| SP116703 | 47 | 15 | 4 | FL_ICGC |
| 03-34157T | 45 | 23 | 4 | FL_GenomeCanada |
| SP194205 | 45 | 12 | 7 | FL_ICGC |
| SP116622 | 44 | 23 | 3 | FL_ICGC |
| SP194234 | 43 | 7 | 8 | DLBCL_ICGC |
| SP59324 | 42 | 4 | 1 | DLBCL_ICGC |
| SP192863 | 41 | 12 | 7 | FL_ICGC |
| FL1005T1 | 40 | 1 | 6 | FL_Kridel |
| HTMCP-01-02-00013-01A-01D | 40 | 40 | NA | DLBCL_HTMCP |
| SP116672 | 40 | 12 | 9 | FL_ICGC |
| HTMCP-01-01-00012-01A-01D | 39 | 39 | NA | DLBCL_HTMCP |
| SP116608 | 39 | 5 | 1 | FL_ICGC |
| SP193300 | 38 | 9 | 4 | DLBCL_ICGC |
| 12-29259T | 37 | 9 | 4 | DLBCL_Gascoyne |
| 14-25416T | 36 | 15 | 3 | FL_GenomeCanada |
| FL2004T1 | 36 | 8 | NA | FL_Kridel |
| SP193993 | 36 | 12 | 6 | FL_ICGC |
| 10-11584_tumorB | 34 | 21 | 6 | DLBCL_LSARP_Trios |
| SP193777 | 34 | 9 | 6 | FL_ICGC |
| HTMCP-01-01-00003-01D-03D | 32 | 32 | NA | DLBCL_HTMCP |
| SP59424 | 32 | 6 | 1 | FL_ICGC |
| FL1001T1 | 31 | 5 | 5 | FL_Kridel |
| HTMCP-01-06-00255-01A-01D | 30 | 30 | NA | DLBCL_HTMCP |
| HTMCP-01-02-00017-01A-01D | 27 | 27 | NA | DLBCL_HTMCP |
| SP59372 | 27 | 5 | 3 | DLBCL_ICGC |
| SP116679 | 26 | 5 | 6 | FL_ICGC |
| SP59376 | 26 | 6 | 5 | DLBCL_ICGC |
| SP59380 | 25 | 6 | 2 | FL_ICGC |
| SP59436 | 24 | 8 | 3 | FL_ICGC |
| 14-33798_tumorA | 23 | 1 | NA | DLBCL_LSARP_Trios |
| HTMCP-01-06-00121-01A-01D | 21 | 21 | NA | DLBCL_HTMCP |
| SP124983 | 17 | 3 | 1 | FL_ICGC |
| HTMCP-01-06-00232-01A-01D | 16 | 16 | NA | DLBCL_HTMCP |
| 14-13938T | 15 | 5 | NA | FL_GenomeCanada |
| HTMCP-01-06-00500-01A-01D | 15 | 15 | NA | DLBCL_HTMCP |
| 04-28140T | 14 | 5 | 1 | DLBCL_Gascoyne |
| HTMCP-01-15-00367-01A-01E | 14 | 14 | NA | DLBCL_HTMCP |
| 10-11584_tumorA | 13 | 6 | 2 | DLBCL_LSARP_Trios |
| HTMCP-01-06-00485-01A-01D | 11 | 11 | NA | DLBCL_HTMCP |
| 05-24561T | 10 | 1 | NA | DLBCL_Marra |
| HTMCP-01-06-00606-01A-01D | 7 | 7 | NA | DLBCL_HTMCP |
| SP193450 | 6 | 4 | 3 | FL_ICGC |
| HTMCP-01-06-00526-01A-01D | 5 | 5 | NA | DLBCL_HTMCP |
| HTMCP-01-07-00336-01A-01E | 2 | 2 | NA | DLBCL_HTMCP |
Note
From this output we can see that there are actually genome-wide mutation calls for a few samples. All these samples are cell lines.
my_meta = dplyr::filter(my_meta,
!cohort %in% "DLBCL_cell_lines")
genome_coding <- get_coding_ssm(
these_samples_metadata = my_meta,
projection = "grch37",
include_silent = TRUE,
this_seq_type = "genome"
)Coding and non-coding mutations
For a high-level overview of what genes the mutations are subset to and their overall mutation incidence in these samples, we can use the GAMBLR.viz function prettyGeneCloud. This function will automatically remove non-coding variants from your data as a convenience feature. We can get around that by assigning the Variant_Classification column for all mutations to imply they are Missense mutations.
fake_maf = mutate(genome_all,Variant_Classification = "Missense_Mutation")
prettyGeneCloud(fake_maf,
zoomout = 0.2,these_genes= unique(genome_all$Hugo_Symbol))
prettyGeneCloud(genome_all,
zoomout = 0.4,these_genes= unique(genome_all$Hugo_Symbol))You will notice that many genes that were prominent in the first cloud are much smaller in the second one. This can be explained by the overwhelming fraction of their mutations representing non-coding variants. This is confirmed by counting up the mutations by Variant_Classification, as demonstrated below.
filter(genome_all,
Hugo_Symbol %in% c("BRINP3","PTPRD","DOCK1","UNC5C")) %>%
group_by(Hugo_Symbol,
Variant_Classification) %>%
count() %>%
kableExtra::kable(format="html")| Hugo_Symbol | Variant_Classification | n |
|---|---|---|
| BRINP3 | 3'Flank | 38 |
| BRINP3 | 3'UTR | 4 |
| BRINP3 | 5'Flank | 38 |
| BRINP3 | 5'UTR | 1 |
| BRINP3 | Frame_Shift_Ins | 1 |
| BRINP3 | Intron | 2247 |
| BRINP3 | Missense_Mutation | 11 |
| BRINP3 | Nonsense_Mutation | 2 |
| BRINP3 | Silent | 5 |
| DOCK1 | 3'Flank | 21 |
| DOCK1 | 3'UTR | 2 |
| DOCK1 | 5'Flank | 17 |
| DOCK1 | Intron | 1630 |
| DOCK1 | Missense_Mutation | 13 |
| DOCK1 | Silent | 6 |
| DOCK1 | Splice_Region | 3 |
| PTPRD | 3'Flank | 16 |
| PTPRD | 3'UTR | 17 |
| PTPRD | 5'Flank | 23 |
| PTPRD | 5'UTR | 3 |
| PTPRD | Intron | 9983 |
| PTPRD | Missense_Mutation | 13 |
| PTPRD | Nonsense_Mutation | 3 |
| PTPRD | Silent | 7 |
| PTPRD | Splice_Region | 2 |
| PTPRD | Splice_Site | 3 |
| UNC5C | 3'Flank | 14 |
| UNC5C | 3'UTR | 20 |
| UNC5C | 5'Flank | 25 |
| UNC5C | Intron | 1613 |
| UNC5C | Missense_Mutation | 10 |
| UNC5C | Silent | 5 |
| UNC5C | Splice_Region | 1 |
aSHM targets
Rather than completely ignoring non-coding variants, we can use this approach to gain an overview of the frequency of mutations in regions that have been identified as targets of aSHM.
# re-run with the cell lines removed
ashm_genome_maf = get_ssm_by_regions(these_samples_metadata = my_meta,
this_seq_type = "genome",
streamlined = F)
prettyGeneCloud(mutate(ashm_genome_maf,
Variant_Classification = "Missense_Mutation"),
zoomout = 0.4,
these_genes= unique(ashm_genome_maf$Hugo_Symbol))
# re-run with the cell lines removed
ashm_genome_streamlined = get_ssm_by_regions(these_samples_metadata = my_meta,
this_seq_type = "genome",
streamlined = TRUE,
use_name_column = TRUE)
#add columns to force prettyGeneCloud to include everything
ashm_genome_streamlined = mutate(ashm_genome_streamlined,
Hugo_Symbol = region_name,
Variant_Classification= "Missense_Mutation",
Tumor_Sample_Barcode = sample_id)
prettyGeneCloud(ashm_genome_streamlined,
zoomout = 0.3,
these_genes= unique(ashm_genome_streamlined$Hugo_Symbol))Summarizing with ggplot2
Word clouds are not useful for communicating the relationship between numeric values. We’ll continue using ggplot2 instead.
ashm_genome_freq = mutate(ashm_genome_streamlined,
gene = str_remove(Hugo_Symbol,"-.+")) %>%
group_by(gene) %>%
summarise(num_mutations=n()) %>%
arrange(desc(num_mutations))
ashm_genome_freq$gene = factor(ashm_genome_freq$gene,
levels = rev(unique(ashm_genome_freq$gene)))
p = ggplot(ashm_genome_freq,aes(y=gene,x=num_mutations)) +
geom_col() +
theme_Morons(base_size=4)
p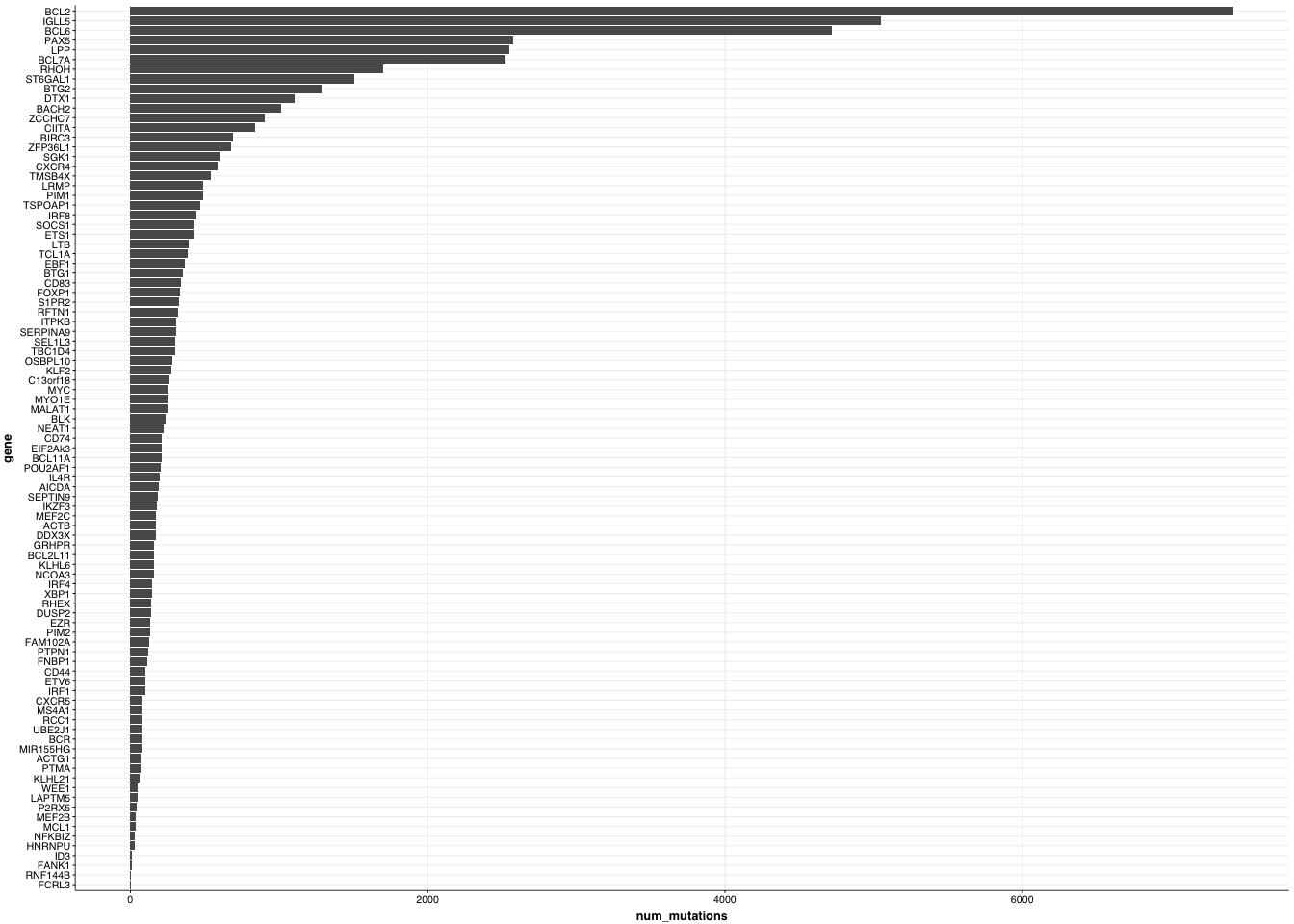
As you can see, the total number of coding + non-coding mutations affecting each of these genes among these samples is quite variable. BCL2, IGLL5, BCL6, PAX5 etc are the most heavily affected.
Building a MAF summary from scratch
Many analyses will probably focus on mutations in protein-coding space and their predicted effect on proteins. For the rest of this tutorial, we’ll delve into this with the mutations we obtained at the start using get_coding_ssm. Here, we’ll work towards reproducing the output of maftools::plotmafSummary, working on one panel at a time.
make_panel1 = function(maf_data,base_size=7,title=""){
vc_counted = maf_data %>%
group_by(Variant_Classification) %>%
count() %>%
arrange(n)
vc_counted$Variant_Classification = factor(
vc_counted$Variant_Classification,
levels=unique(vc_counted$Variant_Classification)
)
mut_cols = get_gambl_colours("mutation")
p1 = ggplot(vc_counted,
aes(x=n,
y=Variant_Classification,
fill=Variant_Classification)) +
geom_col() + scale_fill_manual(values=mut_cols)+
theme_Morons(base_size = base_size,
my_legend_position = "none") +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
) +
ggtitle(title)
p1
}
make_panel1(genome_coding, title="Genomes, coding regions")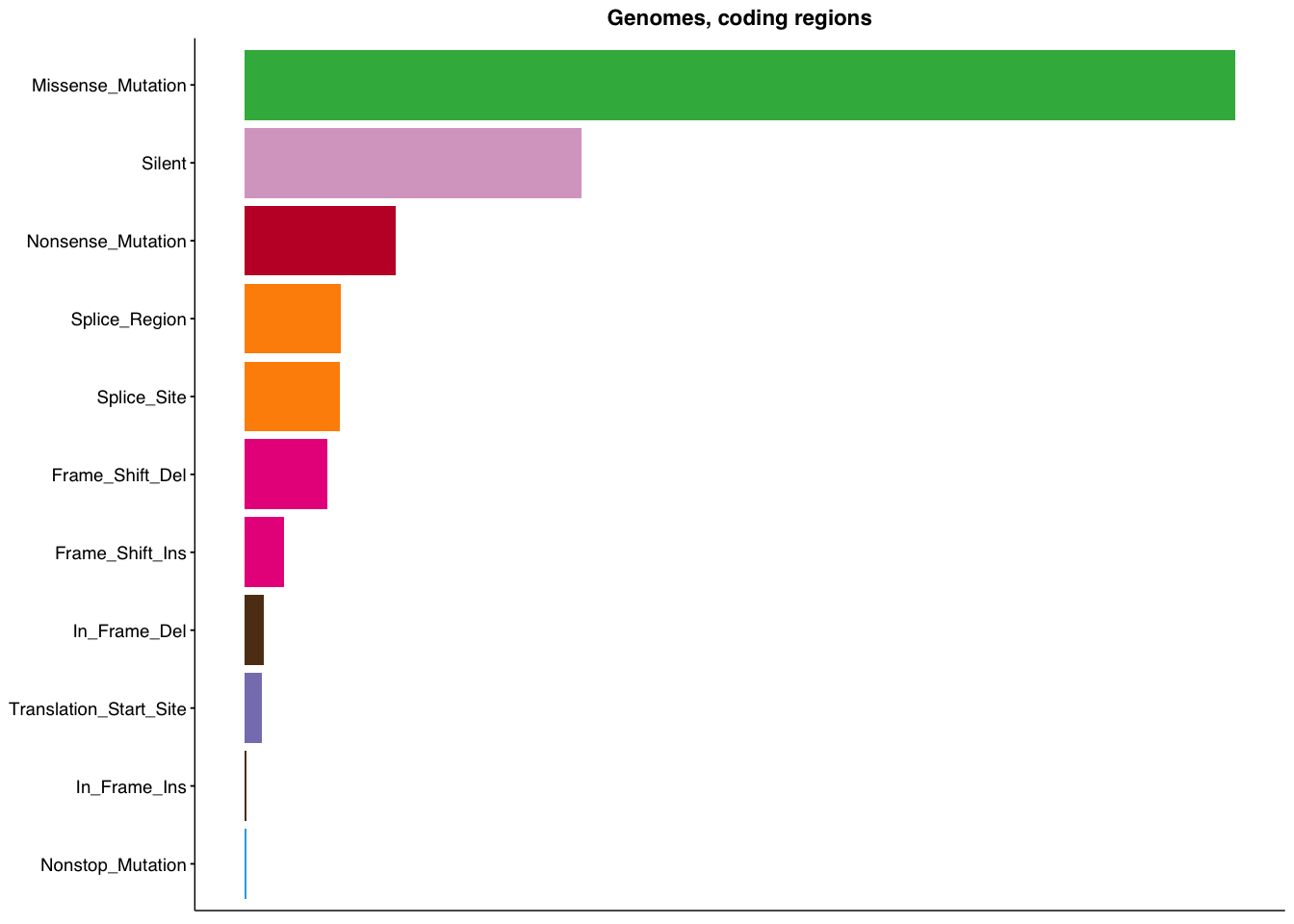
make_panel1(genome_all, title="Genomes, all regions")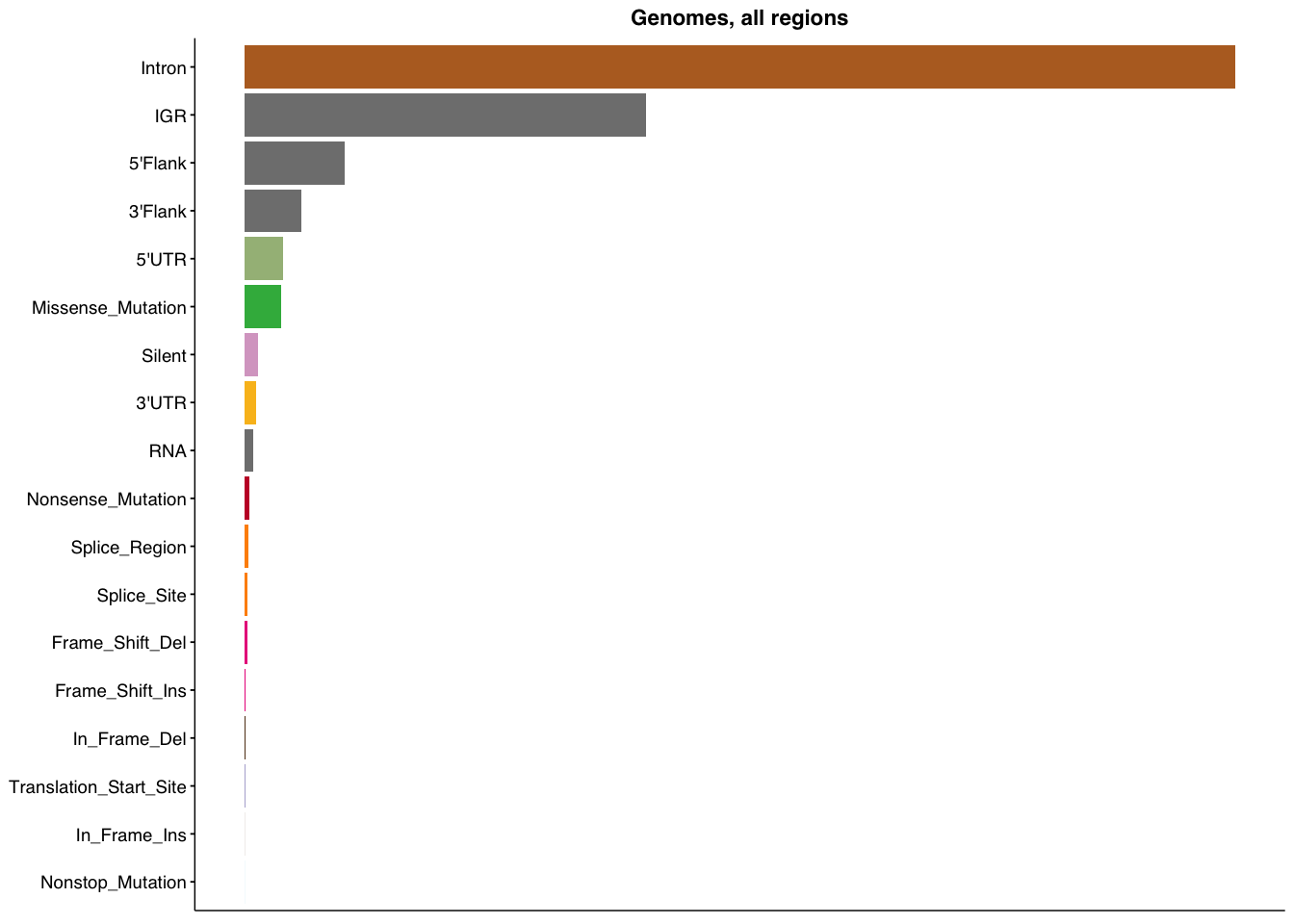
make_panel1(capture_coding, title="Exomes")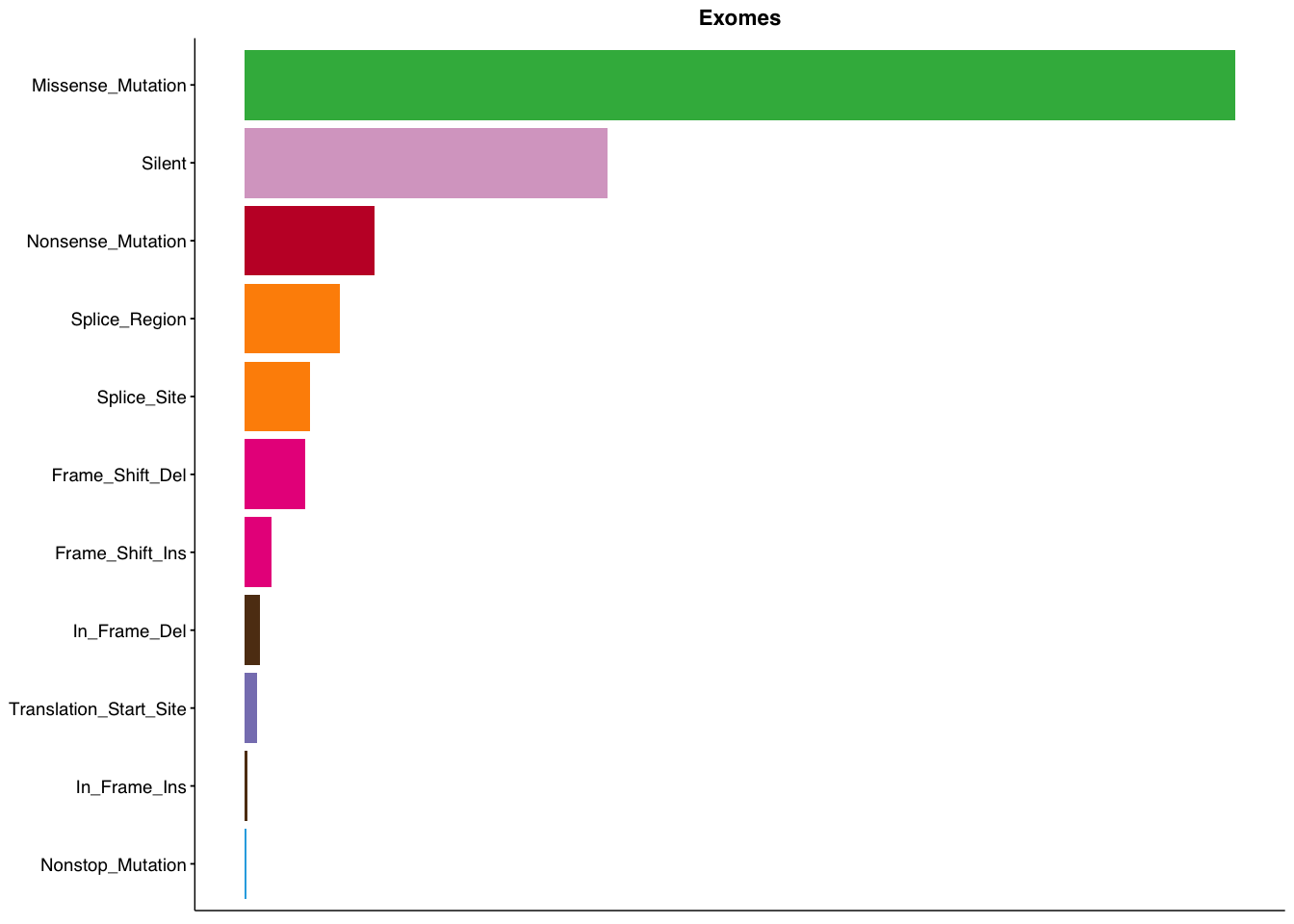
make_panel2 = function(maf_data,base_size=7,title=""){
type_counted = maf_data %>%
group_by(Variant_Type) %>%
count() %>%
arrange(n)
type_counted$Variant_Type = factor(
type_counted$Variant_Type,
levels=unique(type_counted$Variant_Type)
)
mut_cols = c(SNP="purple1",INS="yellow3",DEL="lightblue",DNP="orange","TNP"="lightgreen")
p2 =ggplot(type_counted,aes(x=n,y=Variant_Type,fill=Variant_Type)) +
geom_col() + scale_fill_manual(values=mut_cols)+
theme_Morons(base_size = base_size,my_legend_position = "none") +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
) +
ggtitle(title)
p2
}
make_panel2(genome_coding, title="Genomes, coding regions")
make_panel2(genome_all, title="Genomes, all regions")
make_panel2(capture_coding, title = "Exomes")
make_panel3 = function(maf_data,base_size=7,title=""){
comp = function(base){
chartr("ACTG", "TGAC",base)
}
maf_data = mutate(maf_data,
class = case_when(
Reference_Allele %in% c("T","C") ~
paste0(Reference_Allele,
">",
Tumor_Seq_Allele2),
TRUE ~ paste0(comp(Reference_Allele),
">",
comp(Tumor_Seq_Allele2)))
)
class_counted = maf_data %>% dplyr::filter(Variant_Type == "SNP") %>%
group_by(class) %>% count()
class_counted = mutate(class_counted,class = factor(class,levels=c("C>A","C>G","C>T","T>C","T>A","T>G")))
mut_cols = get_gambl_colours("rainfall")
p3 = ggplot(class_counted,aes(x=n,y=class,fill=class)) +
geom_col() + scale_fill_manual(values=mut_cols)+
theme_Morons(base_size = base_size,my_legend_position = "none") +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
) +
ggtitle(title)
p3
}
make_panel3(genome_coding, title="Genomes, coding regions")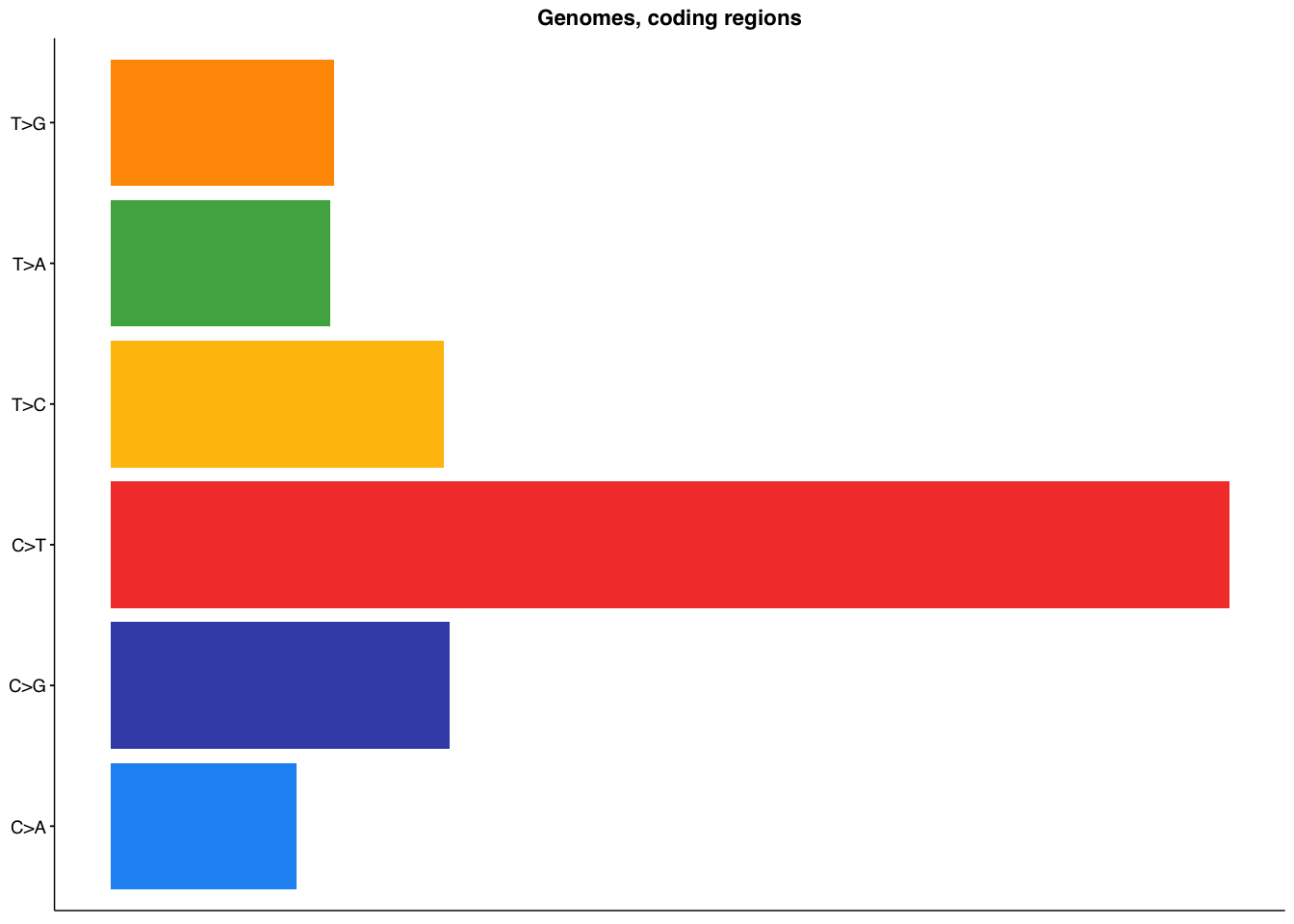
make_panel3(genome_all, title="Genomes, all regions")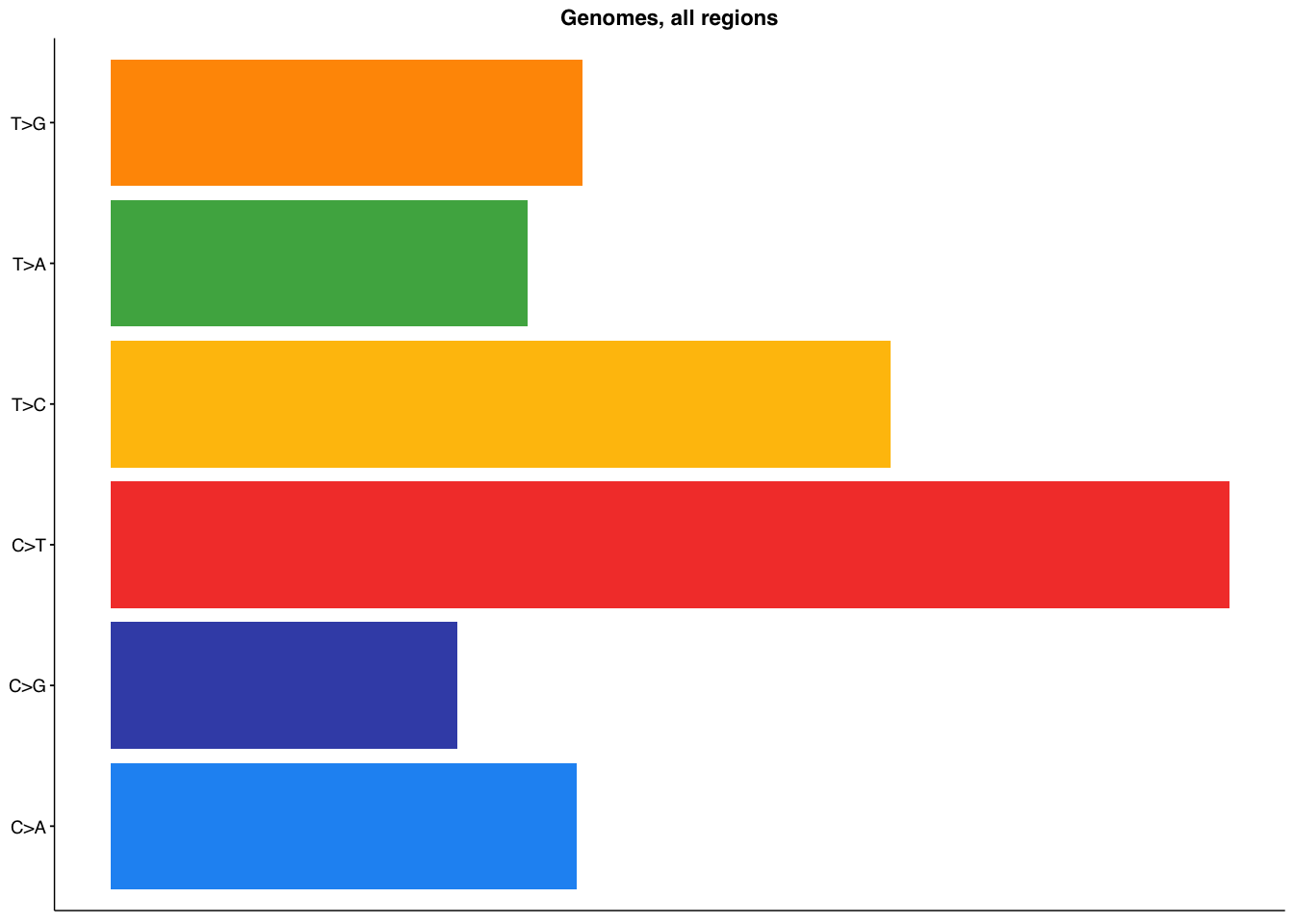
make_panel3(capture_coding, title = "Exomes")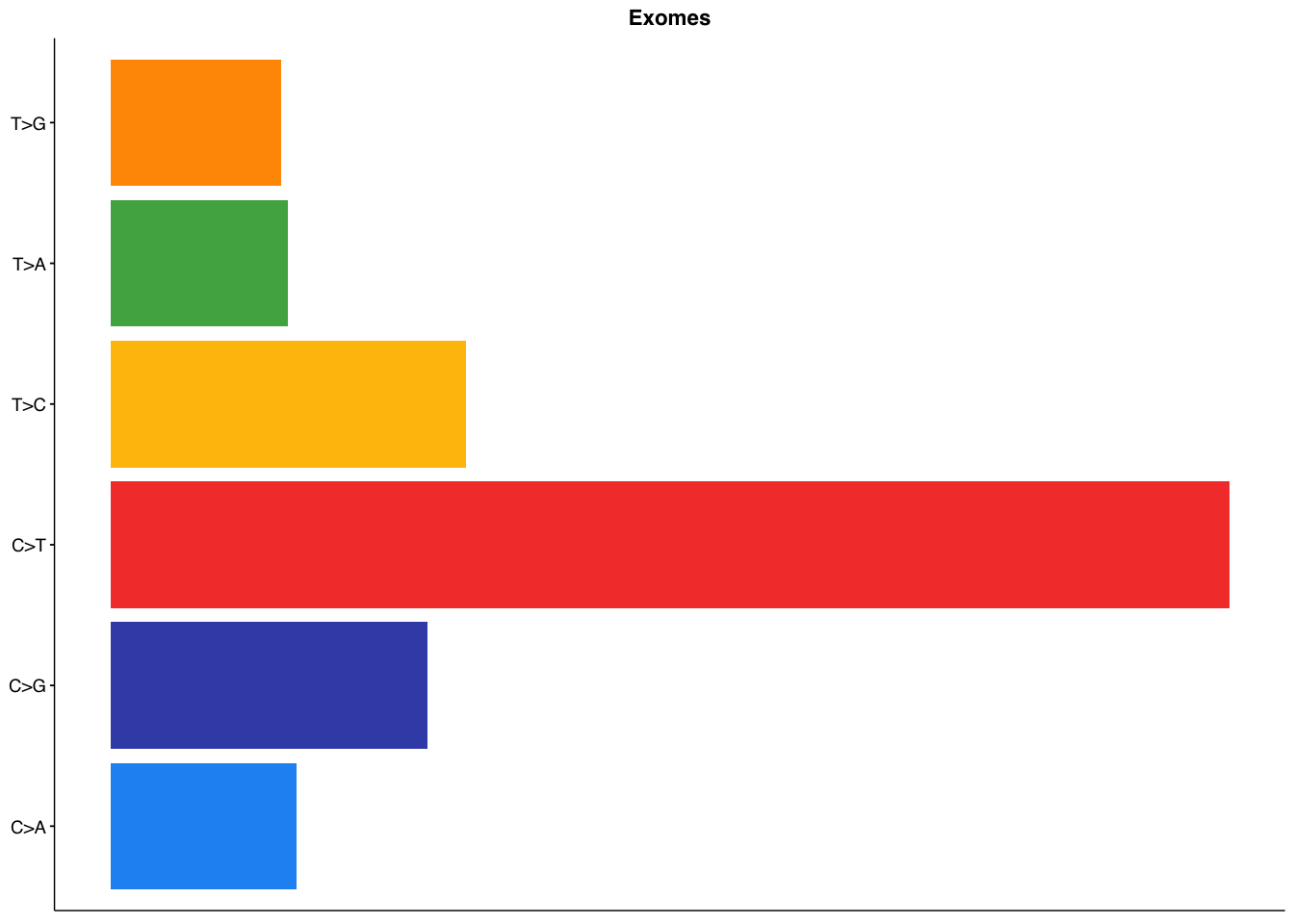
make_panel4 = function(maf_data,base_size=7,title=""){
type_counted = maf_data %>%
group_by(Tumor_Sample_Barcode,Variant_Classification) %>%
count() %>%
arrange(desc(n))
type_counted$Tumor_Sample_Barcode = factor(type_counted$Tumor_Sample_Barcode,
levels=unique(type_counted$Tumor_Sample_Barcode))
mut_cols = get_gambl_colours("mutation")
p4 = ggplot(type_counted,aes(x=Tumor_Sample_Barcode,y=n,fill=Variant_Classification)) +
geom_col() +
scale_fill_manual(values=mut_cols) +
theme_Morons(base_size = base_size,my_legend_position = "none") +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
) +
ggtitle(title)
p4
}
make_panel4(genome_coding, title="Genomes, coding regions")
make_panel4(genome_all, title="Genomes, all regions")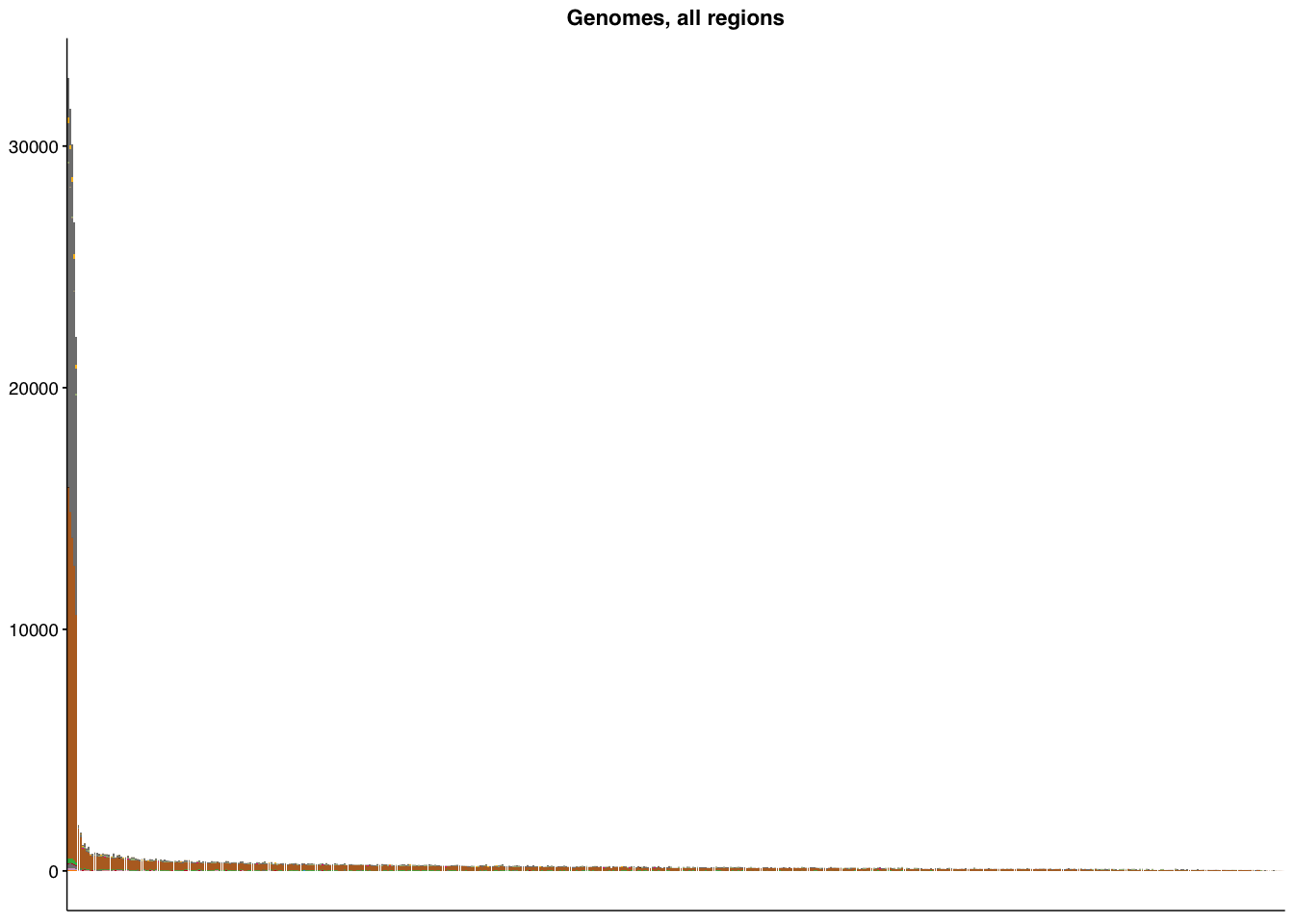
make_panel4(capture_coding, title = "Exomes")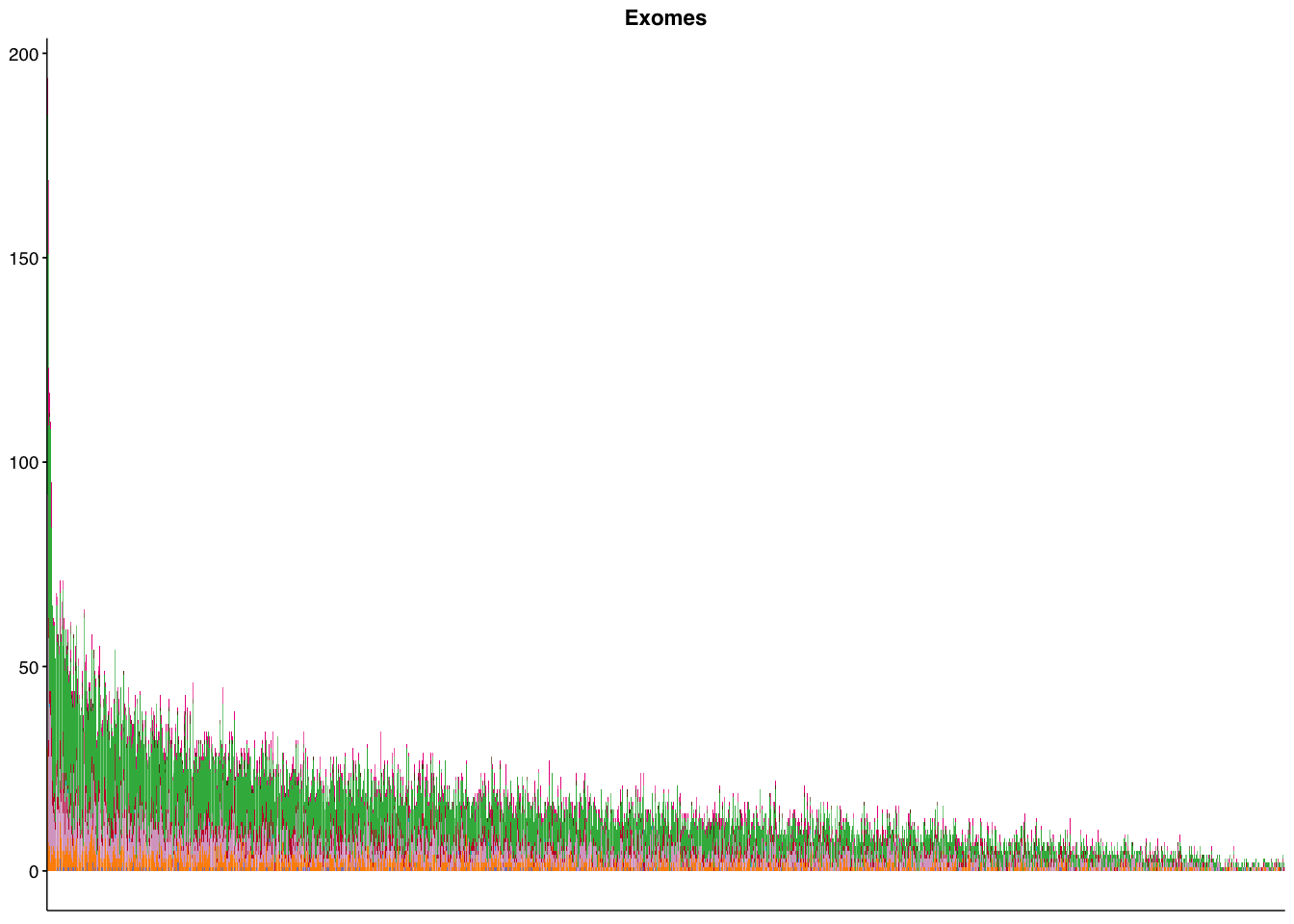
library(ggbeeswarm)
make_panel5 = function(maf_data,base_size=7,point_size=0.5,title=""){
mut_cols = get_gambl_colours()
type_counted = maf_data %>%
group_by(Tumor_Sample_Barcode,Variant_Classification) %>%
count() %>%
arrange(desc(n))
vc_counted = maf_data %>%
group_by(Variant_Classification) %>%
count() %>%
arrange(n)
vc_counted$Variant_Classification = factor(vc_counted$Variant_Classification,
levels=unique(vc_counted$Variant_Classification))
type_counted$Variant_Classification = factor(type_counted$Variant_Classification,
levels=rev(unique(vc_counted$Variant_Classification)))
p5 = ggplot(type_counted,aes(x=Variant_Classification,y=n,colour=Variant_Classification)) +
geom_quasirandom(size=point_size) +
scale_colour_manual(values=mut_cols) +
scale_y_log10() +
theme_Morons(base_size = base_size,my_legend_position = "none") +
theme(axis.title.y =element_blank(),
axis.text.x =element_blank(),
axis.title.x = element_blank(),
axis.ticks.x=element_blank()) +
ggtitle(title)
p5
}
make_panel5(genome_coding, title="Genomes, coding regions")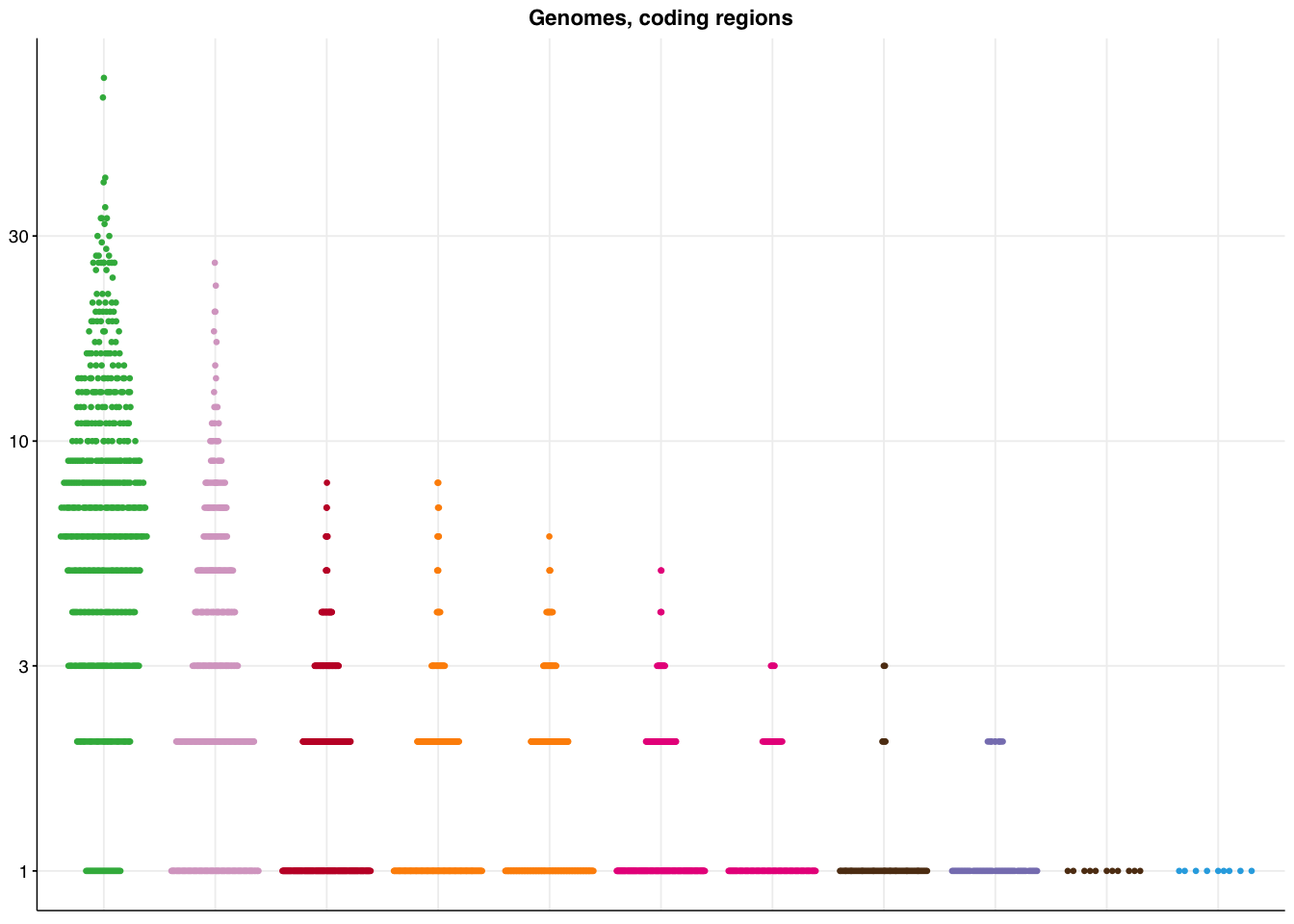
make_panel5(genome_all, title="Genomes, all regions")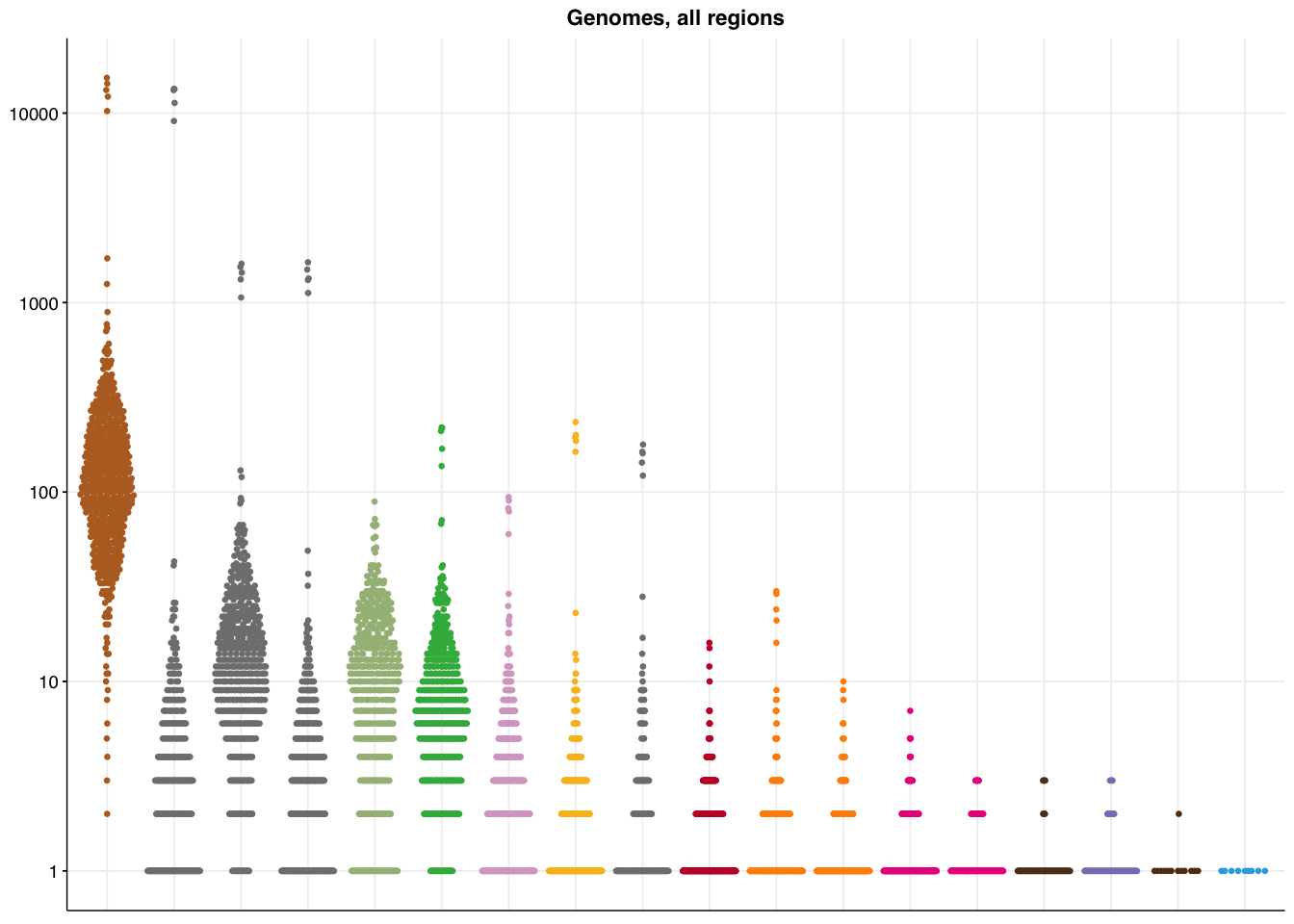
make_panel5(capture_coding, title="Exomes")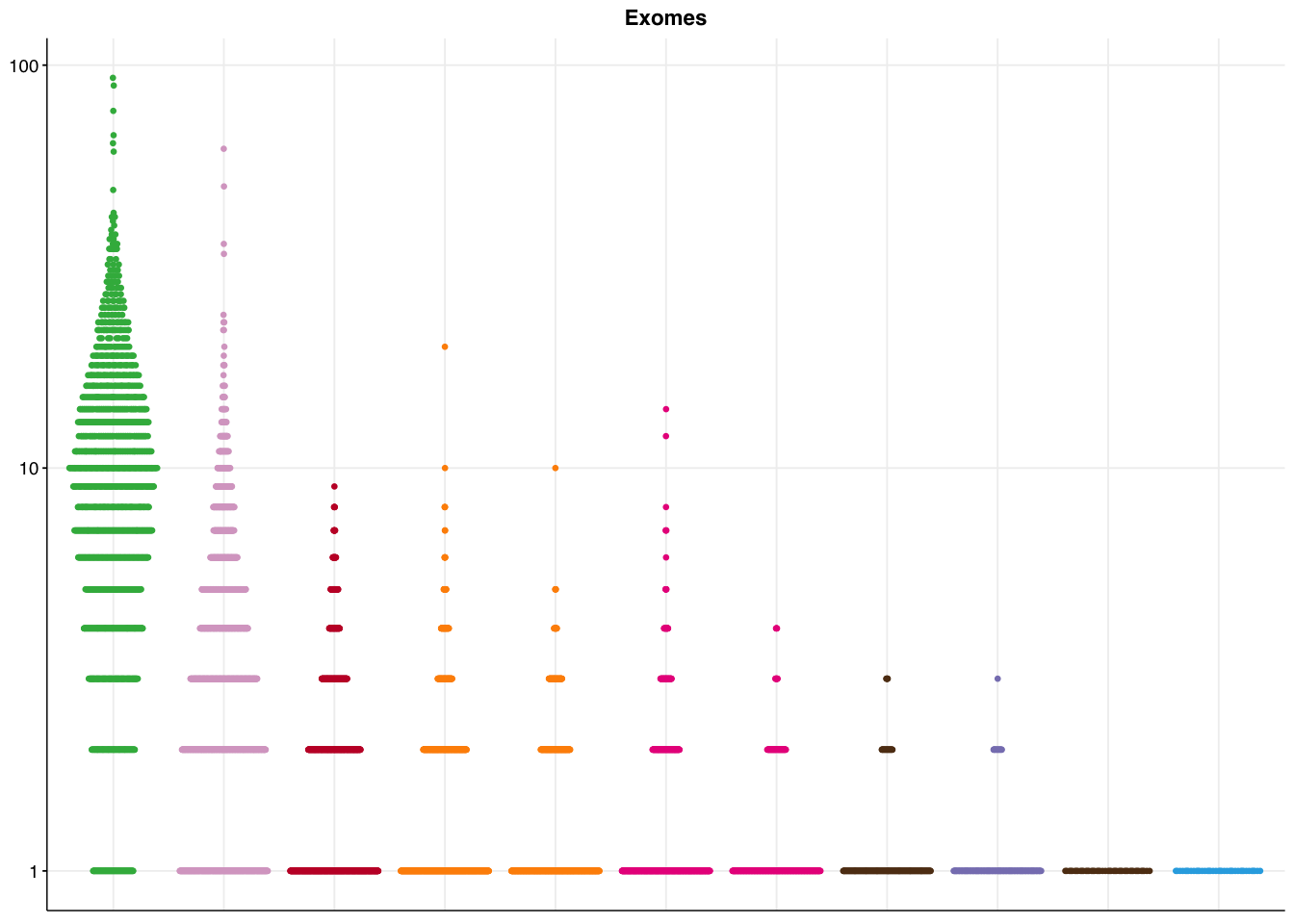
make_panel6 = function(maf_data,base_size=7,top=10,title=""){
type_counted = maf_data %>%
group_by(Hugo_Symbol,Variant_Classification) %>%
count() %>%
arrange(n)
top_n = group_by(type_counted,Hugo_Symbol) %>%
summarise(total=sum(n)) %>%
arrange(desc(total)) %>%
slice_head(n=top) %>%
pull(Hugo_Symbol)
mut_cols = get_gambl_colours()
some_type_counted = dplyr::filter(type_counted,Hugo_Symbol %in% top_n)
some_type_counted$Hugo_Symbol = factor(some_type_counted$Hugo_Symbol,
levels=rev(top_n))
p6 =
ggplot(some_type_counted,aes(y=Hugo_Symbol,x=n,fill=Variant_Classification)) +
geom_col() +
scale_fill_manual(values=mut_cols) +
theme_Morons(base_size = base_size,my_legend_position = "none") +
theme(axis.text.x=element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
axis.ticks.x=element_blank()) +
ggtitle(title)
p6
}
make_panel6(genome_coding,base_size=6, title="Genome, coding regions")
make_panel6(genome_all, title="Genomes, all regions")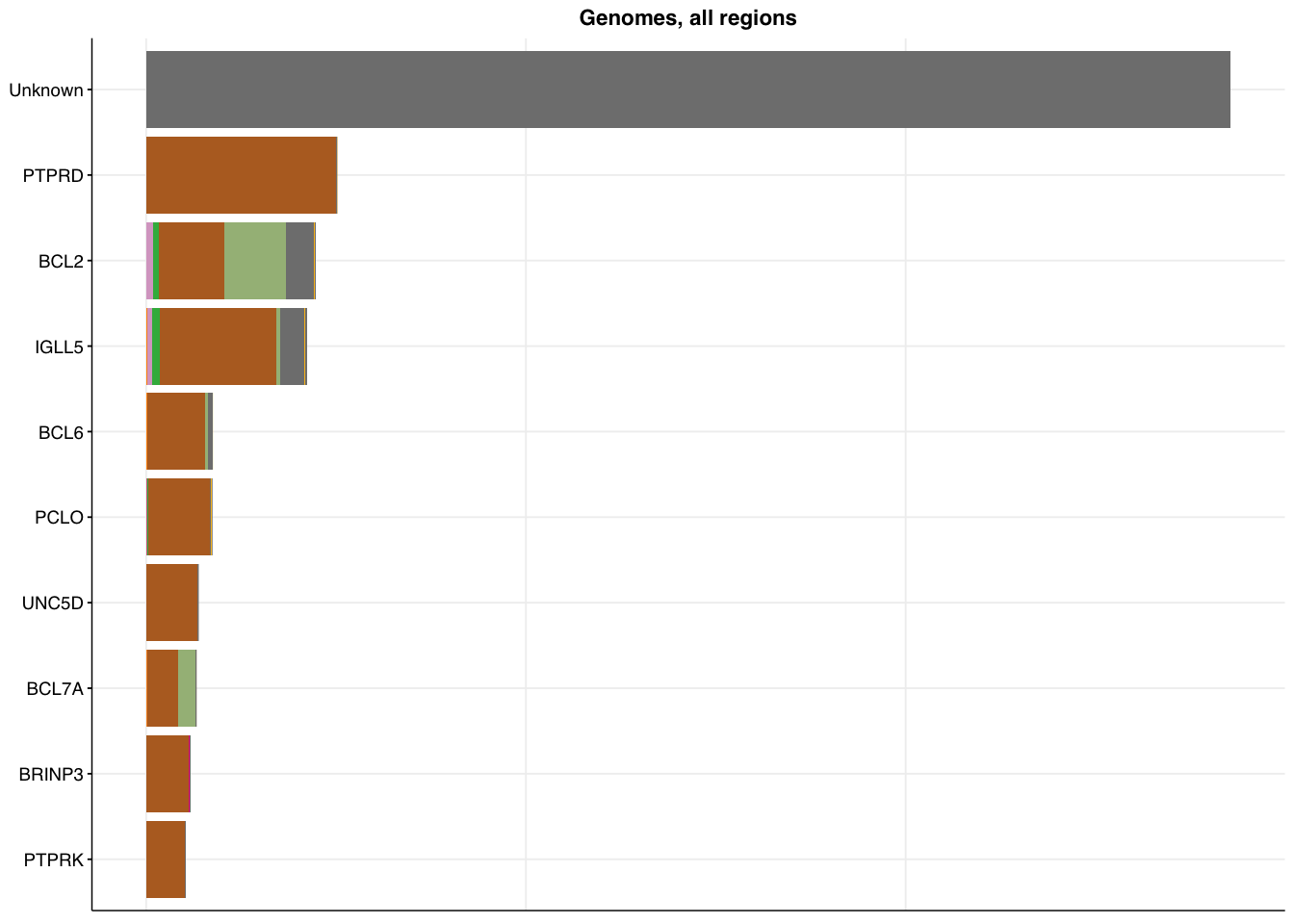
make_panel6(capture_coding, title = "Exomes")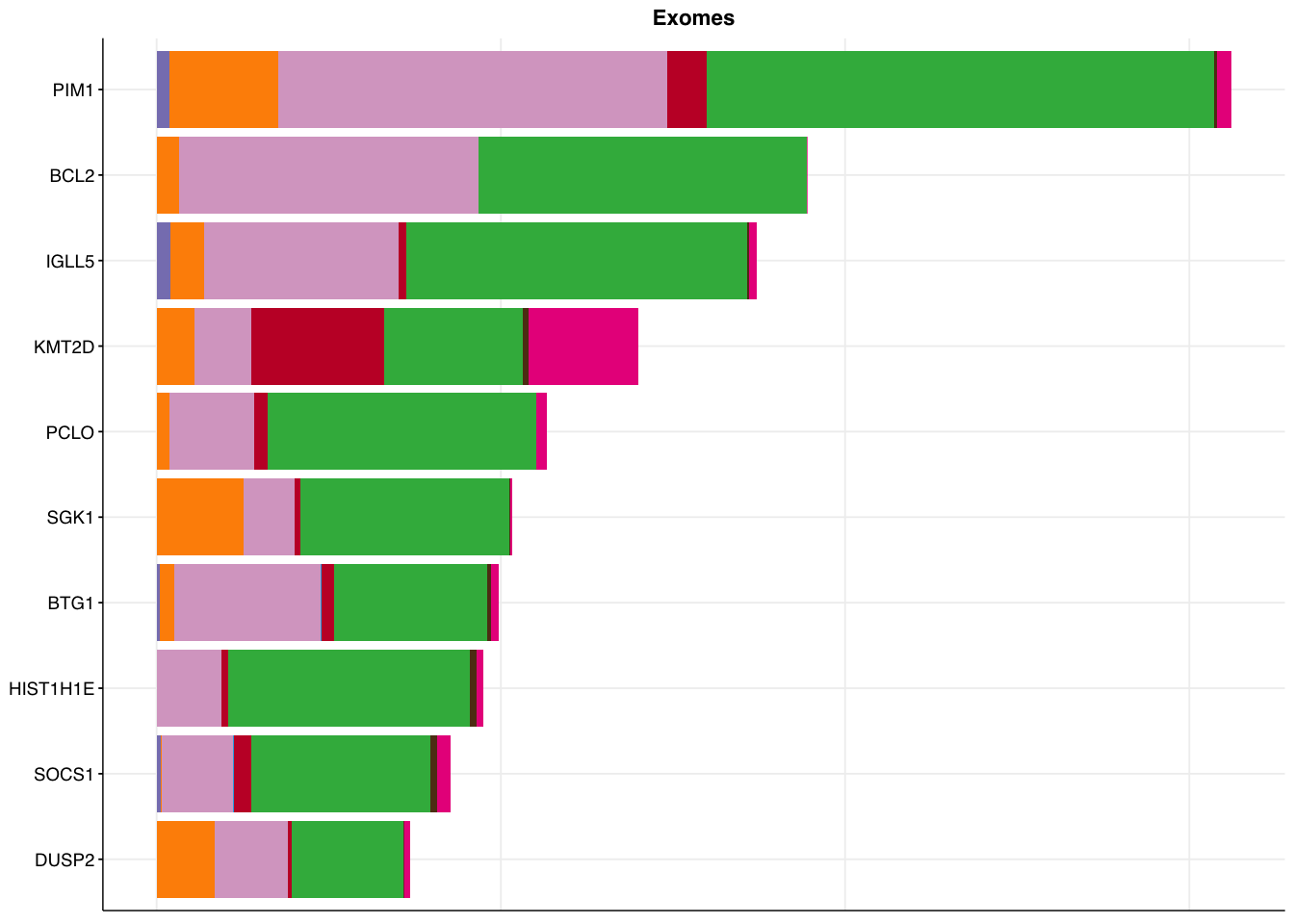
library(cowplot)
bs = 8
ps =0.1
p1 = make_panel1(genome_coding,base_size = bs,title="Variant Classification")
p2 = make_panel2(genome_coding,base_size = bs,title="Variant Type")
p3 = make_panel3(genome_coding,base_size = bs,title="SNV Class")
p4 = make_panel4(genome_coding,base_size = bs,title="Variants per sample")
p5 = make_panel5(genome_coding,base_size = bs,point_size=ps,title="Variant Classification Summary")
p6 = make_panel6(genome_coding,base_size = bs, title="Top 10 genes")
all_p = cowplot::plot_grid(p1,p2,p3,p4,p5,p6,nrow = 2,ncol=3)
all_p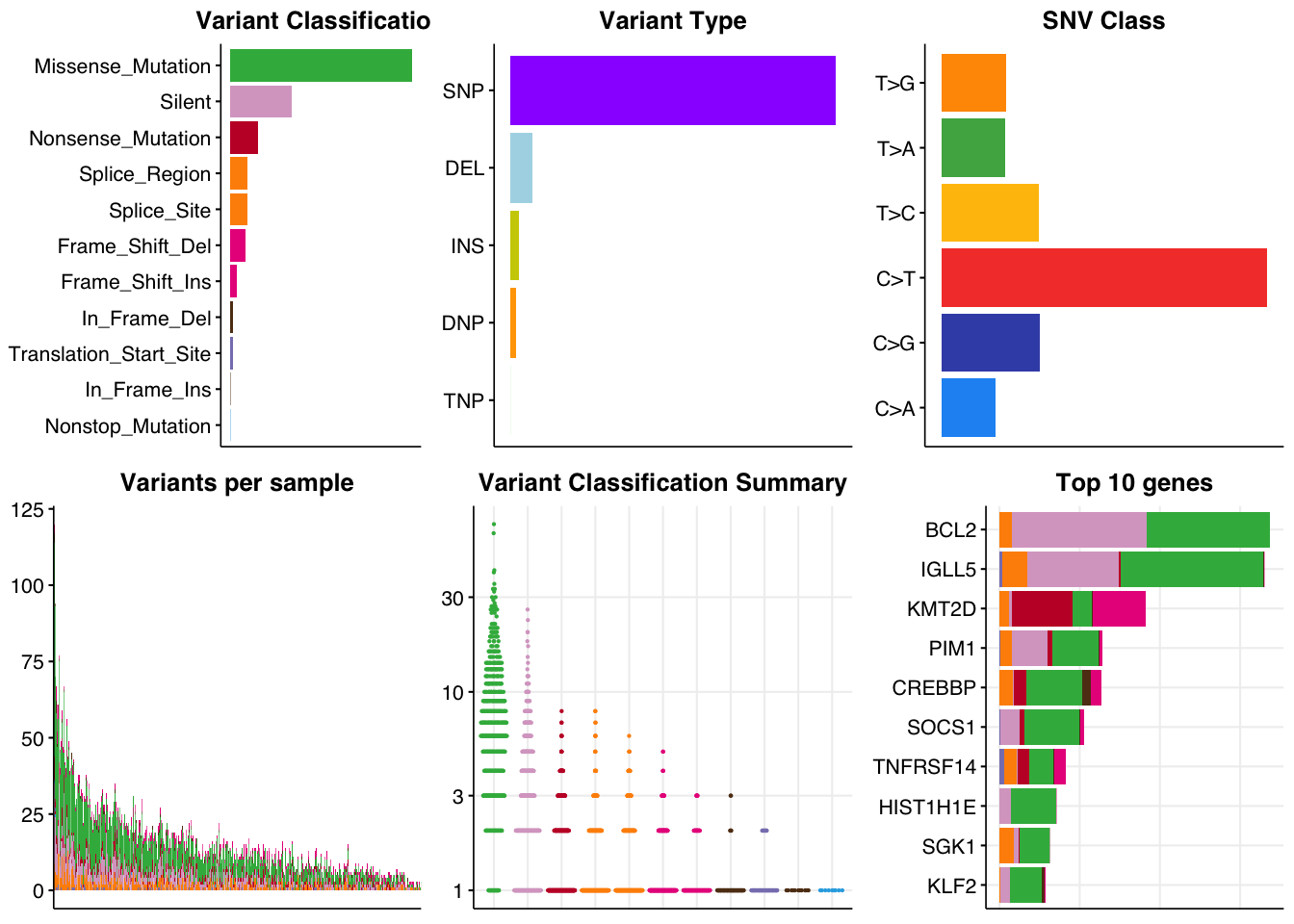
Happy GAMBLing!
/$$$$$$ /$$$$$$ /$$ /$$ /$$$$$$$ /$$ .:::::::
/$$__ $$ /$$__ $$ | $$$ /$$$ | $$__ $$ | $$ .:: .::
| $$ \__/ | $$ \ $$ | $$$$ /$$$$ | $$ \ $$ | $$ .:: .::
| $$ /$$$$ | $$$$$$$$ | $$ $$/$$ $$ | $$$$$$$ | $$ <- .: .::
| $$|_ $$ | $$__ $$ | $$ $$$| $$ | $$__ $$ | $$ .:: .::
| $$ \ $$ | $$ | $$ | $$\ $ | $$ | $$ \ $$ | $$ .:: .::
| $$$$$$/ | $$ | $$ | $$ \/ | $$ | $$$$$$$/ | $$$$$$$$ .:: .::
\______/ |__/ |__/ |__/ |__/ |_______/ |________/
~GENOMIC~~~~~~~~~~~~~OF~~~~~~~~~~~~~~~~~B-CELL~~~~~~~~~~~~~~~~~~IN~~~~~~
~~~~~~~~~~~~ANALYSIS~~~~~~MATURE~~~~~~~~~~~~~~~~~~~LYMPHOMAS~~~~~~~~~~R~