Compare segmented data for multiple samples.
cnvKompare.RdcnvKompare returns a list in variable data formats allowing to evaluate concordance of CNV data between multiple samples.
cnvKompare(
patient_id,
these_sample_ids,
this_seg,
seg_path,
genes_of_interest,
projection = "grch37",
this_seq_type = "genome",
ignore_cytoband_labels = c("acen", "gvar", "stalk"),
max_overlap = 20,
min_concordance = 90,
exclude_sex = FALSE,
return_heatmap = TRUE,
compare_pairwise = TRUE,
show_x_labels = TRUE
)Arguments
- patient_id
Specify patient_id to retrieve sample ids from GAMBL metadata.
- these_sample_ids
Optionally, specify sample ids for comparison.
- this_seg
Optional input data frame of seg file. Must adhere to seg format.
- seg_path
Optionally, specify the path to a local seg file. Must adhere to seg format.
- genes_of_interest
Provide specific genes to be displayed on the time-series plot.
- projection
Argument specifying the projection of seg file, which will determine coordinates of the cytobands. Default is grch37, but hg38 is also accepted.
- this_seq_type
Seq type for returned CN segments. One of "genome" (default) or "capture".
- ignore_cytoband_labels
Cytobands to be ignored. By default, "acen", "gvar", "stalk" are excluded.
- max_overlap
For a time-series plot, how many maximum overlapping points are allowed?
- min_concordance
Integer value from 0 to 100 to indicate the minimum required similarity between cytobands to be considered concordant. The default is 90 (90%).
- exclude_sex
Boolean argument specifying whether to exclude sex chromosomes from calculation. Default is FALSE.
- return_heatmap
Boolean argument specifying whether to return a heatmap of cnvKompare scores. Default is TRUE.
- compare_pairwise
Boolean argument specifying whether to perform pairwise comparisons if there are more than 2 time points in the group. Default is TRUE.
- show_x_labels
Optional boolean parameter for hiding/showing x axis labels, default is TRUE.
Value
A list of overall and pairwise percent concordance, concordant and discordant cytobands, comparison heatmap of cnvKompare scores, and time series ggplot object.
Details
This function will compare CNV data between samples with multiple time points. It can also handle same-sample comparison between different CNV callers if the sample ID is specified in unique fashion. For groups with more than 2 samples, optionally pairwise comparisons can be performed. The comparison is made based on the internally calculated score, which reflects the percentage of each cytoband covered by CNV (rounded to the nearest 5%) and its absolute CN. Optionally, the heatmap of cnvKompare scores can be returned. In addition, the function will return all concordant and discordant cytobands. Finally, the time series plot of CNV log ratios will be returned for all lymphoma genes, with further functionality to subset it to a panel of genes of interest.
Examples
cnvKompare(patient_id = "13-26835",
genes_of_interest = c("EZH2",
"TP53",
"MYC",
"CREBBP",
"GNA13"),
projection = "hg38",
show_x_labels = FALSE)
#> Found 4 samples for patient 13-26835 ...
#> Retreiving the CNV data using GAMBLR ...
#> Calculating CNV concordance ...
#> Building heatmap ...
#> Subsetting lymphoma genes to specified genes of interest ...
#> Performing pairwise comparisons ...
#> DONE!
#> $overall_concordance_pct
#> [1] 12.12
#>
#> $concordant_cytobands
#> # A tibble: 384 × 7
#> ID cb.chromosome cb.start cb.end name score log.ratio
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 13-26835_tumorA chr10 0 3000000 chr10_p15.3 200 0
#> 2 13-26835_tumorB chr10 0 3000000 chr10_p15.3 250 0
#> 3 13-26835_tumorC chr10 0 3000000 chr10_p15.3 200 0
#> 4 13-26835_tumorD chr10 0 3000000 chr10_p15.3 200 0
#> 5 13-26835_tumorA chr10 41600000 45500000 chr10_q11.… 200 0
#> 6 13-26835_tumorB chr10 41600000 45500000 chr10_q11.… 250 0
#> 7 13-26835_tumorC chr10 41600000 45500000 chr10_q11.… 200 0
#> 8 13-26835_tumorD chr10 41600000 45500000 chr10_q11.… 200 0
#> 9 13-26835_tumorA chr10 128800000 133797422 chr10_q26.3 200 0
#> 10 13-26835_tumorB chr10 128800000 133797422 chr10_q26.3 250 0.585
#> # ℹ 374 more rows
#>
#> $discordant_cytobands
#> # A tibble: 2,784 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0 200
#> 2 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0.585 300
#> 3 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 -0.180 200
#> 4 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0 200
#> 5 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0 200
#> 6 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0.585 300
#> 7 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 -0.180 200
#> 8 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0 200
#> 9 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0 200
#> 10 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0.585 300
#> # ℹ 2,774 more rows
#>
#> $Heatmap
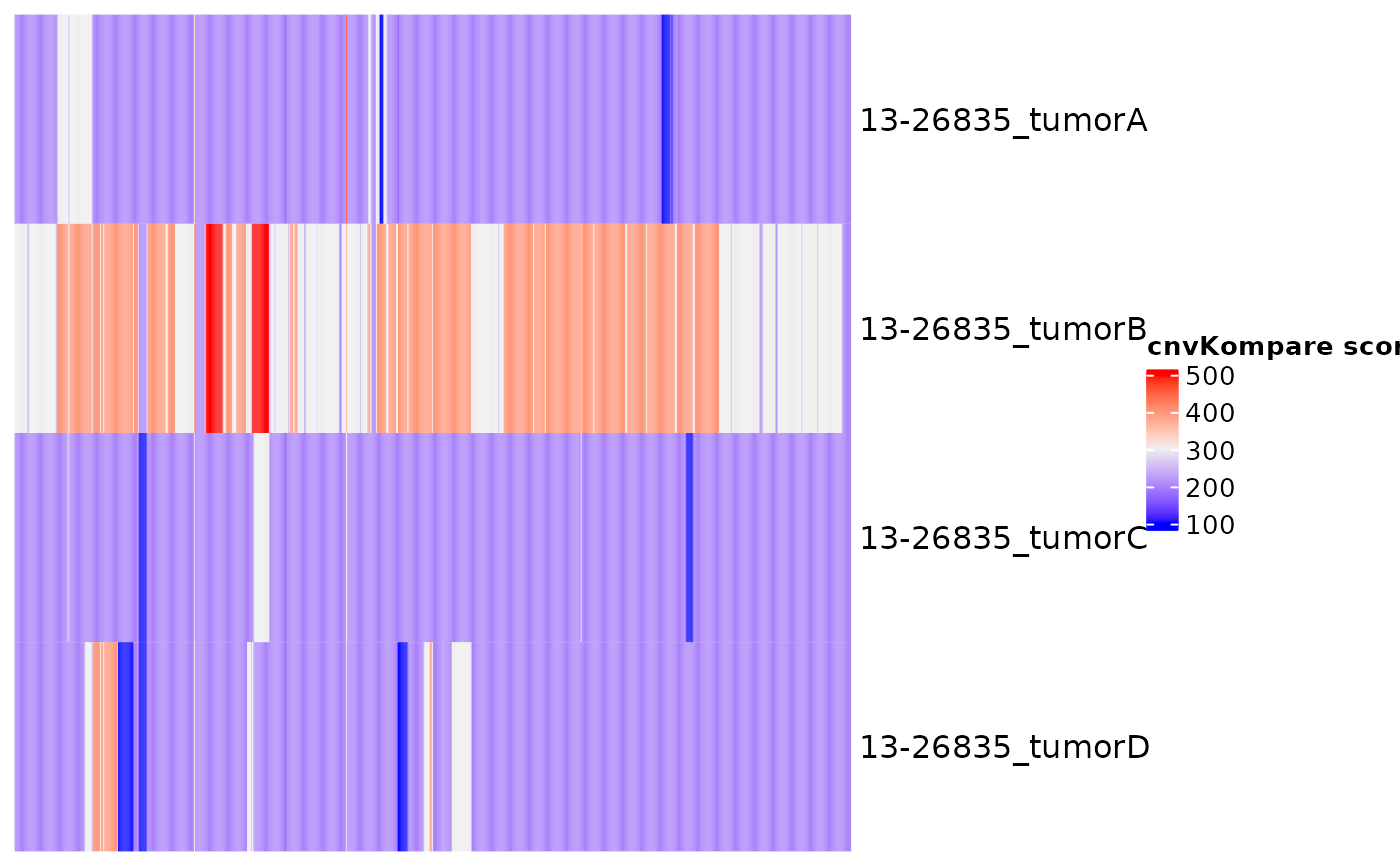 #>
#> $time_plot
#>
#> $time_plot
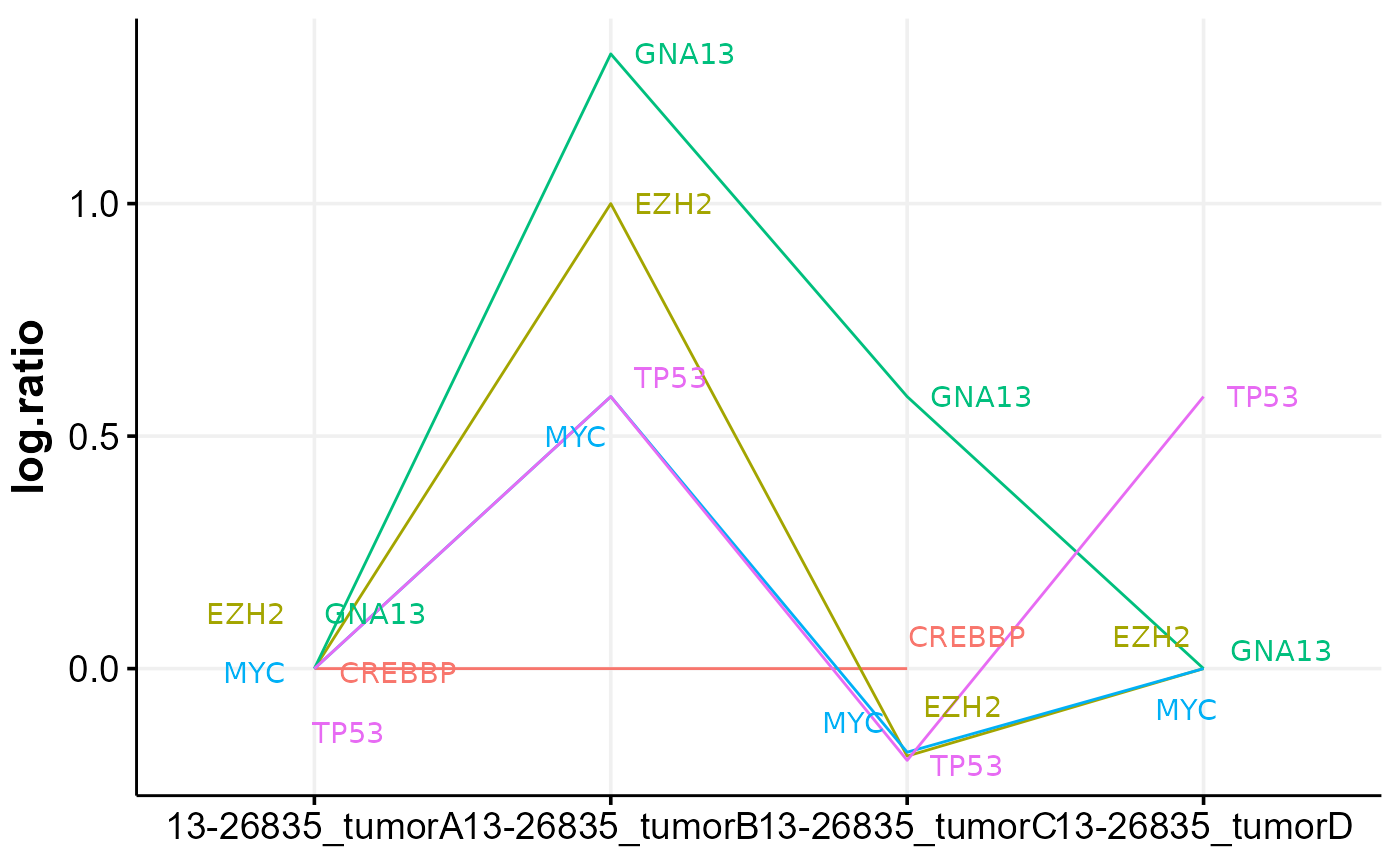 #>
#> $pairwise_comparisons
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorB`
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorB`$pairwise_concordance_pct
#> [1] 4.17
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorB`$concordant_cytobands
#> # A tibble: 66 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorA chr13 31600000 33400000 chr13_q13.1 200
#> 2 13-26835_tumorB chr13 31600000 33400000 chr13_q13.1 200
#> 3 13-26835_tumorA chr13 33400000 34900000 chr13_q13.2 200
#> 4 13-26835_tumorB chr13 33400000 34900000 chr13_q13.2 200
#> 5 13-26835_tumorA chr13 34900000 39500000 chr13_q13.3 200
#> 6 13-26835_tumorB chr13 34900000 39500000 chr13_q13.3 200
#> 7 13-26835_tumorA chr13 39500000 44600000 chr13_q14.11 200
#> 8 13-26835_tumorB chr13 39500000 44600000 chr13_q14.11 200
#> 9 13-26835_tumorA chr13 44600000 45200000 chr13_q14.12 200
#> 10 13-26835_tumorB chr13 44600000 45200000 chr13_q14.12 200
#> # ℹ 56 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorB`$discordant_cytobands
#> # A tibble: 1,518 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0 200
#> 2 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0.585 300
#> 3 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0 200
#> 4 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0.585 300
#> 5 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0 200
#> 6 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0.585 300
#> 7 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0 200
#> 8 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0.585 300
#> 9 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0 200
#> 10 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0.585 300
#> # ℹ 1,508 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorC`
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorC`$pairwise_concordance_pct
#> [1] 88.13
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorC`$concordant_cytobands
#> # A tibble: 1,396 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorA chr10 34200000 38000000 chr10_p11.21 200
#> 2 13-26835_tumorC chr10 34200000 38000000 chr10_p11.21 200
#> 3 13-26835_tumorA chr10 31100000 34200000 chr10_p11.22 200
#> 4 13-26835_tumorC chr10 31100000 34200000 chr10_p11.22 200
#> 5 13-26835_tumorA chr10 29300000 31100000 chr10_p11.23 200
#> 6 13-26835_tumorC chr10 29300000 31100000 chr10_p11.23 200
#> 7 13-26835_tumorA chr10 24300000 29300000 chr10_p12.1 200
#> 8 13-26835_tumorC chr10 24300000 29300000 chr10_p12.1 200
#> 9 13-26835_tumorA chr10 22300000 24300000 chr10_p12.2 200
#> 10 13-26835_tumorC chr10 22300000 24300000 chr10_p12.2 200
#> # ℹ 1,386 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorC`$discordant_cytobands
#> # A tibble: 188 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr11 48800000 5.10e7 chr1… 100 0.585 250
#> 2 13-26835_tum… chr11 48800000 5.10e7 chr1… 100 -0.219 200
#> 3 13-26835_tum… chr11 43400000 4.88e7 chr1… 100 0.585 300
#> 4 13-26835_tum… chr11 43400000 4.88e7 chr1… 100 -0.219 200
#> 5 13-26835_tum… chr11 36400000 4.34e7 chr1… 100 0.585 300
#> 6 13-26835_tum… chr11 36400000 4.34e7 chr1… 100 -0.219 200
#> 7 13-26835_tum… chr11 31000000 3.64e7 chr1… 100 0.585 300
#> 8 13-26835_tum… chr11 31000000 3.64e7 chr1… 100 -0.219 200
#> 9 13-26835_tum… chr11 27200000 3.10e7 chr1… 100 0.585 300
#> 10 13-26835_tum… chr11 27200000 3.10e7 chr1… 100 -0.219 200
#> # ℹ 178 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorD`
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorD`$pairwise_concordance_pct
#> [1] 81.44
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorD`$concordant_cytobands
#> # A tibble: 1,290 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorA chr10 34200000 38000000 chr10_p11.21 200
#> 2 13-26835_tumorD chr10 34200000 38000000 chr10_p11.21 200
#> 3 13-26835_tumorA chr10 31100000 34200000 chr10_p11.22 200
#> 4 13-26835_tumorD chr10 31100000 34200000 chr10_p11.22 200
#> 5 13-26835_tumorA chr10 29300000 31100000 chr10_p11.23 200
#> 6 13-26835_tumorD chr10 29300000 31100000 chr10_p11.23 200
#> 7 13-26835_tumorA chr10 24300000 29300000 chr10_p12.1 200
#> 8 13-26835_tumorD chr10 24300000 29300000 chr10_p12.1 200
#> 9 13-26835_tumorA chr10 22300000 24300000 chr10_p12.2 200
#> 10 13-26835_tumorD chr10 22300000 24300000 chr10_p12.2 200
#> # ℹ 1,280 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorD`$discordant_cytobands
#> # A tibble: 294 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr11 48800000 5.10e7 chr1… 100 0.585 250
#> 2 13-26835_tum… chr11 48800000 5.10e7 chr1… 100 0 200
#> 3 13-26835_tum… chr11 43400000 4.88e7 chr1… 100 0.585 300
#> 4 13-26835_tum… chr11 43400000 4.88e7 chr1… 100 0 200
#> 5 13-26835_tum… chr11 36400000 4.34e7 chr1… 100 0.585 300
#> 6 13-26835_tum… chr11 36400000 4.34e7 chr1… 100 0 200
#> 7 13-26835_tum… chr11 31000000 3.64e7 chr1… 100 0.585 300
#> 8 13-26835_tum… chr11 31000000 3.64e7 chr1… 100 0 200
#> 9 13-26835_tum… chr11 27200000 3.10e7 chr1… 100 0.585 300
#> 10 13-26835_tum… chr11 27200000 3.10e7 chr1… 100 0 200
#> # ℹ 284 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorC`
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorC`$pairwise_concordance_pct
#> [1] 3.16
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorC`$concordant_cytobands
#> # A tibble: 50 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorB chr15 20500000 25500000 chr15_q11.2 200
#> 2 13-26835_tumorC chr15 20500000 25500000 chr15_q11.2 200
#> 3 13-26835_tumorB chr15 25500000 27800000 chr15_q12 200
#> 4 13-26835_tumorC chr15 25500000 27800000 chr15_q12 200
#> 5 13-26835_tumorB chr15 27800000 30000000 chr15_q13.1 200
#> 6 13-26835_tumorC chr15 27800000 30000000 chr15_q13.1 200
#> 7 13-26835_tumorB chr15 30000000 30900000 chr15_q13.2 200
#> 8 13-26835_tumorC chr15 30000000 30900000 chr15_q13.2 200
#> 9 13-26835_tumorB chr15 30900000 33400000 chr15_q13.3 200
#> 10 13-26835_tumorC chr15 30900000 33400000 chr15_q13.3 200
#> # ℹ 40 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorC`$discordant_cytobands
#> # A tibble: 1,534 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0.585 300
#> 2 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 -0.180 200
#> 3 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0.585 300
#> 4 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 -0.180 200
#> 5 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0.585 300
#> 6 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 -0.180 200
#> 7 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0.585 300
#> 8 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 -0.180 200
#> 9 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0.585 300
#> 10 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 -0.180 200
#> # ℹ 1,524 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorD`
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorD`$pairwise_concordance_pct
#> [1] 7.07
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorD`$concordant_cytobands
#> # A tibble: 112 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorB chr12 30500000 33200000 chr12_p11.21 400
#> 2 13-26835_tumorD chr12 30500000 33200000 chr12_p11.21 400
#> 3 13-26835_tumorB chr12 27600000 30500000 chr12_p11.22 400
#> 4 13-26835_tumorD chr12 27600000 30500000 chr12_p11.22 400
#> 5 13-26835_tumorB chr12 26300000 27600000 chr12_p11.23 400
#> 6 13-26835_tumorD chr12 26300000 27600000 chr12_p11.23 400
#> 7 13-26835_tumorB chr12 21100000 26300000 chr12_p12.1 400
#> 8 13-26835_tumorD chr12 21100000 26300000 chr12_p12.1 400
#> 9 13-26835_tumorB chr12 19800000 21100000 chr12_p12.2 400
#> 10 13-26835_tumorD chr12 19800000 21100000 chr12_p12.2 400
#> # ℹ 102 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorD`$discordant_cytobands
#> # A tibble: 1,472 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0.585 300
#> 2 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0 200
#> 3 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0.585 300
#> 4 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0 200
#> 5 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0.585 300
#> 6 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0 200
#> 7 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0.585 300
#> 8 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0 200
#> 9 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0.585 300
#> 10 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0 200
#> # ℹ 1,462 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorC--13-26835_tumorD`
#> $pairwise_comparisons$`13-26835_tumorC--13-26835_tumorD`$pairwise_concordance_pct
#> [1] 85.1
#>
#> $pairwise_comparisons$`13-26835_tumorC--13-26835_tumorD`$concordant_cytobands
#> # A tibble: 1,348 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorC chr10 34200000 38000000 chr10_p11.21 200
#> 2 13-26835_tumorD chr10 34200000 38000000 chr10_p11.21 200
#> 3 13-26835_tumorC chr10 31100000 34200000 chr10_p11.22 200
#> 4 13-26835_tumorD chr10 31100000 34200000 chr10_p11.22 200
#> 5 13-26835_tumorC chr10 29300000 31100000 chr10_p11.23 200
#> 6 13-26835_tumorD chr10 29300000 31100000 chr10_p11.23 200
#> 7 13-26835_tumorC chr10 24300000 29300000 chr10_p12.1 200
#> 8 13-26835_tumorD chr10 24300000 29300000 chr10_p12.1 200
#> 9 13-26835_tumorC chr10 22300000 24300000 chr10_p12.2 200
#> 10 13-26835_tumorD chr10 22300000 24300000 chr10_p12.2 200
#> # ℹ 1,338 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorC--13-26835_tumorD`$discordant_cytobands
#> # A tibble: 236 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr11 2.8 e6 1.17e7 chr1… 100 0.585 233.
#> 2 13-26835_tum… chr11 2.8 e6 1.17e7 chr1… 100 0 200
#> 3 13-26835_tum… chr11 0 2.8 e6 chr1… 100 0 250
#> 4 13-26835_tum… chr11 0 2.8 e6 chr1… 100 0 200
#> 5 13-26835_tum… chr11 1.03e8 1.11e8 chr1… 100 -0.180 200
#> 6 13-26835_tum… chr11 1.03e8 1.11e8 chr1… 100 0 233.
#> 7 13-26835_tum… chr11 1.11e8 1.13e8 chr1… 100 -0.180 200
#> 8 13-26835_tum… chr11 1.11e8 1.13e8 chr1… 100 0.585 300
#> 9 13-26835_tum… chr11 1.13e8 1.15e8 chr1… 100 -0.180 200
#> 10 13-26835_tum… chr11 1.13e8 1.15e8 chr1… 100 0.585 300
#> # ℹ 226 more rows
#>
#>
#>
#>
#> $pairwise_comparisons
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorB`
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorB`$pairwise_concordance_pct
#> [1] 4.17
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorB`$concordant_cytobands
#> # A tibble: 66 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorA chr13 31600000 33400000 chr13_q13.1 200
#> 2 13-26835_tumorB chr13 31600000 33400000 chr13_q13.1 200
#> 3 13-26835_tumorA chr13 33400000 34900000 chr13_q13.2 200
#> 4 13-26835_tumorB chr13 33400000 34900000 chr13_q13.2 200
#> 5 13-26835_tumorA chr13 34900000 39500000 chr13_q13.3 200
#> 6 13-26835_tumorB chr13 34900000 39500000 chr13_q13.3 200
#> 7 13-26835_tumorA chr13 39500000 44600000 chr13_q14.11 200
#> 8 13-26835_tumorB chr13 39500000 44600000 chr13_q14.11 200
#> 9 13-26835_tumorA chr13 44600000 45200000 chr13_q14.12 200
#> 10 13-26835_tumorB chr13 44600000 45200000 chr13_q14.12 200
#> # ℹ 56 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorB`$discordant_cytobands
#> # A tibble: 1,518 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0 200
#> 2 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0.585 300
#> 3 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0 200
#> 4 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0.585 300
#> 5 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0 200
#> 6 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0.585 300
#> 7 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0 200
#> 8 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0.585 300
#> 9 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0 200
#> 10 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0.585 300
#> # ℹ 1,508 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorC`
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorC`$pairwise_concordance_pct
#> [1] 88.13
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorC`$concordant_cytobands
#> # A tibble: 1,396 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorA chr10 34200000 38000000 chr10_p11.21 200
#> 2 13-26835_tumorC chr10 34200000 38000000 chr10_p11.21 200
#> 3 13-26835_tumorA chr10 31100000 34200000 chr10_p11.22 200
#> 4 13-26835_tumorC chr10 31100000 34200000 chr10_p11.22 200
#> 5 13-26835_tumorA chr10 29300000 31100000 chr10_p11.23 200
#> 6 13-26835_tumorC chr10 29300000 31100000 chr10_p11.23 200
#> 7 13-26835_tumorA chr10 24300000 29300000 chr10_p12.1 200
#> 8 13-26835_tumorC chr10 24300000 29300000 chr10_p12.1 200
#> 9 13-26835_tumorA chr10 22300000 24300000 chr10_p12.2 200
#> 10 13-26835_tumorC chr10 22300000 24300000 chr10_p12.2 200
#> # ℹ 1,386 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorC`$discordant_cytobands
#> # A tibble: 188 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr11 48800000 5.10e7 chr1… 100 0.585 250
#> 2 13-26835_tum… chr11 48800000 5.10e7 chr1… 100 -0.219 200
#> 3 13-26835_tum… chr11 43400000 4.88e7 chr1… 100 0.585 300
#> 4 13-26835_tum… chr11 43400000 4.88e7 chr1… 100 -0.219 200
#> 5 13-26835_tum… chr11 36400000 4.34e7 chr1… 100 0.585 300
#> 6 13-26835_tum… chr11 36400000 4.34e7 chr1… 100 -0.219 200
#> 7 13-26835_tum… chr11 31000000 3.64e7 chr1… 100 0.585 300
#> 8 13-26835_tum… chr11 31000000 3.64e7 chr1… 100 -0.219 200
#> 9 13-26835_tum… chr11 27200000 3.10e7 chr1… 100 0.585 300
#> 10 13-26835_tum… chr11 27200000 3.10e7 chr1… 100 -0.219 200
#> # ℹ 178 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorD`
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorD`$pairwise_concordance_pct
#> [1] 81.44
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorD`$concordant_cytobands
#> # A tibble: 1,290 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorA chr10 34200000 38000000 chr10_p11.21 200
#> 2 13-26835_tumorD chr10 34200000 38000000 chr10_p11.21 200
#> 3 13-26835_tumorA chr10 31100000 34200000 chr10_p11.22 200
#> 4 13-26835_tumorD chr10 31100000 34200000 chr10_p11.22 200
#> 5 13-26835_tumorA chr10 29300000 31100000 chr10_p11.23 200
#> 6 13-26835_tumorD chr10 29300000 31100000 chr10_p11.23 200
#> 7 13-26835_tumorA chr10 24300000 29300000 chr10_p12.1 200
#> 8 13-26835_tumorD chr10 24300000 29300000 chr10_p12.1 200
#> 9 13-26835_tumorA chr10 22300000 24300000 chr10_p12.2 200
#> 10 13-26835_tumorD chr10 22300000 24300000 chr10_p12.2 200
#> # ℹ 1,280 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorA--13-26835_tumorD`$discordant_cytobands
#> # A tibble: 294 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr11 48800000 5.10e7 chr1… 100 0.585 250
#> 2 13-26835_tum… chr11 48800000 5.10e7 chr1… 100 0 200
#> 3 13-26835_tum… chr11 43400000 4.88e7 chr1… 100 0.585 300
#> 4 13-26835_tum… chr11 43400000 4.88e7 chr1… 100 0 200
#> 5 13-26835_tum… chr11 36400000 4.34e7 chr1… 100 0.585 300
#> 6 13-26835_tum… chr11 36400000 4.34e7 chr1… 100 0 200
#> 7 13-26835_tum… chr11 31000000 3.64e7 chr1… 100 0.585 300
#> 8 13-26835_tum… chr11 31000000 3.64e7 chr1… 100 0 200
#> 9 13-26835_tum… chr11 27200000 3.10e7 chr1… 100 0.585 300
#> 10 13-26835_tum… chr11 27200000 3.10e7 chr1… 100 0 200
#> # ℹ 284 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorC`
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorC`$pairwise_concordance_pct
#> [1] 3.16
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorC`$concordant_cytobands
#> # A tibble: 50 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorB chr15 20500000 25500000 chr15_q11.2 200
#> 2 13-26835_tumorC chr15 20500000 25500000 chr15_q11.2 200
#> 3 13-26835_tumorB chr15 25500000 27800000 chr15_q12 200
#> 4 13-26835_tumorC chr15 25500000 27800000 chr15_q12 200
#> 5 13-26835_tumorB chr15 27800000 30000000 chr15_q13.1 200
#> 6 13-26835_tumorC chr15 27800000 30000000 chr15_q13.1 200
#> 7 13-26835_tumorB chr15 30000000 30900000 chr15_q13.2 200
#> 8 13-26835_tumorC chr15 30000000 30900000 chr15_q13.2 200
#> 9 13-26835_tumorB chr15 30900000 33400000 chr15_q13.3 200
#> 10 13-26835_tumorC chr15 30900000 33400000 chr15_q13.3 200
#> # ℹ 40 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorC`$discordant_cytobands
#> # A tibble: 1,534 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0.585 300
#> 2 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 -0.180 200
#> 3 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0.585 300
#> 4 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 -0.180 200
#> 5 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0.585 300
#> 6 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 -0.180 200
#> 7 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0.585 300
#> 8 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 -0.180 200
#> 9 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0.585 300
#> 10 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 -0.180 200
#> # ℹ 1,524 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorD`
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorD`$pairwise_concordance_pct
#> [1] 7.07
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorD`$concordant_cytobands
#> # A tibble: 112 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorB chr12 30500000 33200000 chr12_p11.21 400
#> 2 13-26835_tumorD chr12 30500000 33200000 chr12_p11.21 400
#> 3 13-26835_tumorB chr12 27600000 30500000 chr12_p11.22 400
#> 4 13-26835_tumorD chr12 27600000 30500000 chr12_p11.22 400
#> 5 13-26835_tumorB chr12 26300000 27600000 chr12_p11.23 400
#> 6 13-26835_tumorD chr12 26300000 27600000 chr12_p11.23 400
#> 7 13-26835_tumorB chr12 21100000 26300000 chr12_p12.1 400
#> 8 13-26835_tumorD chr12 21100000 26300000 chr12_p12.1 400
#> 9 13-26835_tumorB chr12 19800000 21100000 chr12_p12.2 400
#> 10 13-26835_tumorD chr12 19800000 21100000 chr12_p12.2 400
#> # ℹ 102 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorB--13-26835_tumorD`$discordant_cytobands
#> # A tibble: 1,472 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0.585 300
#> 2 13-26835_tum… chr10 34200000 3.8 e7 chr1… 100 0 200
#> 3 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0.585 300
#> 4 13-26835_tum… chr10 31100000 3.42e7 chr1… 100 0 200
#> 5 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0.585 300
#> 6 13-26835_tum… chr10 29300000 3.11e7 chr1… 100 0 200
#> 7 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0.585 300
#> 8 13-26835_tum… chr10 24300000 2.93e7 chr1… 100 0 200
#> 9 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0.585 300
#> 10 13-26835_tum… chr10 22300000 2.43e7 chr1… 100 0 200
#> # ℹ 1,462 more rows
#>
#>
#> $pairwise_comparisons$`13-26835_tumorC--13-26835_tumorD`
#> $pairwise_comparisons$`13-26835_tumorC--13-26835_tumorD`$pairwise_concordance_pct
#> [1] 85.1
#>
#> $pairwise_comparisons$`13-26835_tumorC--13-26835_tumorD`$concordant_cytobands
#> # A tibble: 1,348 × 6
#> ID cb.chromosome cb.start cb.end name score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl>
#> 1 13-26835_tumorC chr10 34200000 38000000 chr10_p11.21 200
#> 2 13-26835_tumorD chr10 34200000 38000000 chr10_p11.21 200
#> 3 13-26835_tumorC chr10 31100000 34200000 chr10_p11.22 200
#> 4 13-26835_tumorD chr10 31100000 34200000 chr10_p11.22 200
#> 5 13-26835_tumorC chr10 29300000 31100000 chr10_p11.23 200
#> 6 13-26835_tumorD chr10 29300000 31100000 chr10_p11.23 200
#> 7 13-26835_tumorC chr10 24300000 29300000 chr10_p12.1 200
#> 8 13-26835_tumorD chr10 24300000 29300000 chr10_p12.1 200
#> 9 13-26835_tumorC chr10 22300000 24300000 chr10_p12.2 200
#> 10 13-26835_tumorD chr10 22300000 24300000 chr10_p12.2 200
#> # ℹ 1,338 more rows
#>
#> $pairwise_comparisons$`13-26835_tumorC--13-26835_tumorD`$discordant_cytobands
#> # A tibble: 236 × 8
#> ID cb.chromosome cb.start cb.end name pct_covered log.ratio score
#> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 13-26835_tum… chr11 2.8 e6 1.17e7 chr1… 100 0.585 233.
#> 2 13-26835_tum… chr11 2.8 e6 1.17e7 chr1… 100 0 200
#> 3 13-26835_tum… chr11 0 2.8 e6 chr1… 100 0 250
#> 4 13-26835_tum… chr11 0 2.8 e6 chr1… 100 0 200
#> 5 13-26835_tum… chr11 1.03e8 1.11e8 chr1… 100 -0.180 200
#> 6 13-26835_tum… chr11 1.03e8 1.11e8 chr1… 100 0 233.
#> 7 13-26835_tum… chr11 1.11e8 1.13e8 chr1… 100 -0.180 200
#> 8 13-26835_tum… chr11 1.11e8 1.13e8 chr1… 100 0.585 300
#> 9 13-26835_tum… chr11 1.13e8 1.15e8 chr1… 100 -0.180 200
#> 10 13-26835_tum… chr11 1.13e8 1.15e8 chr1… 100 0.585 300
#> # ℹ 226 more rows
#>
#>
#>