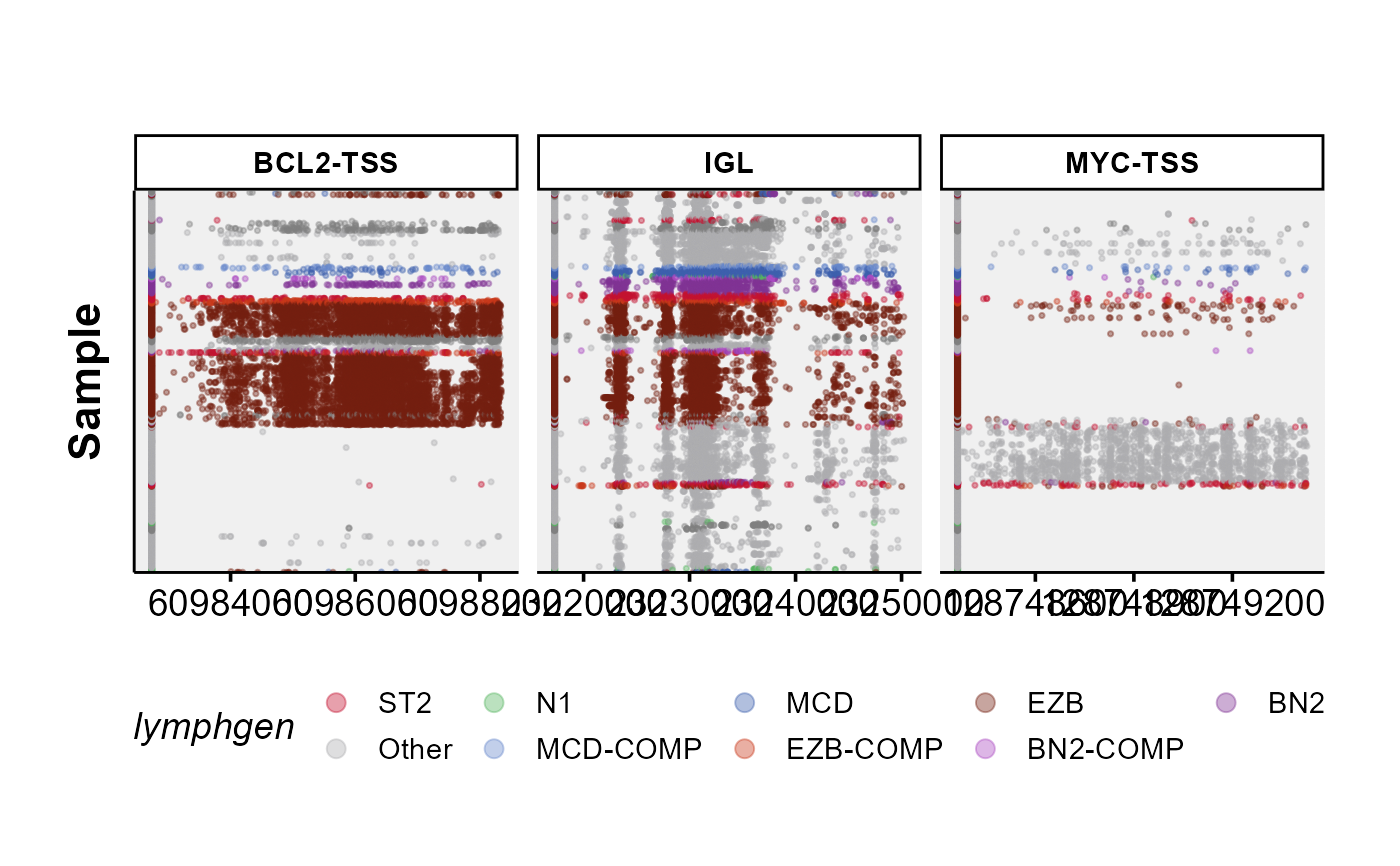
ASHM Multi-panel Rainbow Plot
ashm_multi_rainbow_plot.RdGenerates a colourful multi-panel overview of hypermutation in regions of interest across many samples.
ashm_multi_rainbow_plot(
regions_bed,
regions_to_display,
exclude_classifications,
metadata,
seq_type,
custom_colours,
classification_column = "lymphgen",
maf_data,
verbose = TRUE
)Arguments
- regions_bed
Bed file with chromosome coordinates, should contain columns chr, start, end, name (with these exact names).
- regions_to_display
Optional vector of names from default regions_bed to use.
- exclude_classifications
Optional argument for excluding specific classifications from a metadeta file.
- metadata
A metadata file already subsetted and arranged on the order you want the samples vertically displayed.
- seq_type
the seqtype you want results back for if
maf_datais not provided.- custom_colours
Provide named vector (or named list of vectors) containing custom annotation colours if you do not want to use standartized pallette.
- classification_column
Optional. Override default column for assigning the labels used for colouring in the figure.
- maf_data
An already loaded maf, if no provided, this function will call
get_ssm_by_region, using the regions supplied intoregions_bed.- verbose
Set to FALSE to rpevent printing the full regions bed file to the console. Default is TRUE.
Value
Nothing
Details
The input for this function is a bed-file with the following columns; chr, start, end, name.
Note that for this function to work, the column names must be exactly this.
The user also needs to specify a vector of names (regions_to_display) to further control what regions are to be displayed on the returned plot.
It is also possible to exclude specific classifications from the metadata file. This is achieved with exclude_classifications.
In addition the user can also use the metadata parameter to use an already subset and arranged metadata table.
This function will call get_ssm_by_region if maf_data is not called. For more info, refer to the parameter descriptions of this function.
Examples
#get lymphgen colours
lymphgen_colours = get_gambl_colours(classification = "lymphgen")
#build plot
ashm_multi_rainbow_plot(regions_to_display = c("BCL2-TSS",
"MYC-TSS",
"SGK1-TSS",
"IGL"),
custom_colours = lymphgen_colours,
seq_type = "genome")
#> chr_name hg19_start hg19_end gene region
#> 1 chr1 6661482 6662702 KLHL21 TSS
#> 2 chr1 23885584 23885835 ID3 TSS
#> 3 chr1 150550814 150552135 MCL1 intronic
#> 4 chr1 203274698 203275778 BTG2 intronic
#> 5 chr1 226864857 226873452 ITPKB intronic
#> 6 chr1 226920563 226927885 ITPKB TSS
#> 7 chr1 226921088 226927982 ITPKB intron-1
#> 8 chr2 60773789 60783486 BCL11A TSS
#> 9 chr2 96808901 96811913 DUSP2 intron-1
#> 10 chr2 111874854 111882174 BCL2L11 TSS
#> 11 chr2 136874728 136875461 CXCR4 intronic
#> 12 chr3 16546433 16556786 RFTN1 TSS
#> 13 chr3 32020518 32024930 OSBPL10 TSS-1
#> 14 chr3 71104986 71119518 FOXP1 TSS-4
#> 15 chr3 71176427 71180960 FOXP1 TSS-3
#> 16 chr3 71350633 71357665 FOXP1 TSS-2
#> 17 chr3 71503103 71504870 FOXP1 intron-1b
#> 18 chr3 71550753 71551478 FOXP1 intron-1a
#> 19 chr3 71626341 71635648 FOXP1 TSS-1
#> 20 chr3 101546669 101547704 NFKBIZ TSS-1
#> 21 chr3 101568239 101569274 NFKBIZ TSS-2
#> 22 chr3 183269360 183274139 KLHL6 TSS
#> 23 chr3 186739628 186740875 ST6GAL1 TSS-1
#> 24 chr3 186782522 186784438 ST6GAL1 intronic-1
#> 25 chr3 187458526 187464632 BCL6 TSS
#> 26 chr3 187771678 187801626 LPP TSS-1
#> 27 chr3 187801627 187813568 LPP TSS-2
#> 28 chr3 187813569 187955254 LPP TSS-3
#> 29 chr3 187955255 187969220 LPP TSS-4
#> 30 chr3 188377178 188401951 LPP intronic-1
#> 31 chr3 188401952 188461879 LPP intronic-2
#> 32 chr3 188461880 188474683 LPP intronic-3
#> 33 chr3 188474684 188491248 LPP intronic-4
#> 34 chr4 40193105 40204231 RHOH TSS
#> 35 chr5 88131209 88174373 MEF2C TSS-3
#> 36 chr5 88174374 88182243 MEF2C TSS-2
#> 37 chr5 88182244 88206620 MEF2C TSS-1
#> 38 chr5 131823933 131826458 IRF1 TSS
#> 39 chr5 149790977 149792349 CD74 TSS
#> 40 chr5 158500476 158532769 EBF1 TSS
#> 41 chr6 390572 394093 IRF4 TSS
#> 42 chr6 14118026 14120025 CD83 TSS
#> 43 chr6 31548325 31550717 LTB intron-1
#> 44 chr6 37138104 37139804 PIM1 TSS
#> 45 chr6 90981034 91016134 BACH2 TSS
#> 46 chr6 134487960 134499859 SGK1 TSS-1
#> 47 chr6 159237903 159240216 EZR TSS
#> 48 chr7 5568297 5570856 ACTB TSS
#> 49 chr8 11347723 11355318 BLK TSS
#> 50 chr8 128748352 128749427 MYC TSS
#> 51 chr9 37023396 37027663 PAX5 intron-1
#> 52 chr9 37029849 37037154 PAX5 TSS-1
#> 53 chr9 37192080 37207549 ZCCHC7 intron-4
#> 54 chr9 37275952 37306152 ZCCHC7 intron-2
#> 55 chr9 37312655 37328260 ZCCHC7 intron-3
#> 56 chr9 37329706 37340398 ZCCHC7 intron-1
#> 57 chr9 37369209 37372160 PAX5 distal-enhancer-1
#> 58 chr9 37382267 37385854 PAX5 distal-enhancer-2
#> 59 chr9 37395932 37409239 PAX5 distal-enhancer-3
#> 60 chr9 37423010 37425279 GRHPR TSS
#> 61 chr10 127578912 127591133 FANK1 TSS
#> 62 chr11 9595246 9599502 WEE1 intronic
#> 63 chr11 35156769 35164248 CD44 TSS
#> 64 chr11 60223385 60225310 MS4A1 TSS
#> 65 chr11 65190369 65192380 NEAT1 ncRNA
#> 66 chr11 65265237 65268359 MALAT1 ncRNA
#> 67 chr11 102188170 102190077 BIRC3 TSS
#> 68 chr11 111248078 111250224 POU2AF1 TSS
#> 69 chr11 128339774 128345731 ETS1 introns
#> 70 chr11 128388492 128394163 ETS1 TSS-2
#> 71 chr12 11796001 11812968 ETV6 TSS
#> 72 chr12 25204045 25213569 LRMP TSS
#> 73 chr12 92537999 92539598 BTG1 TSS
#> 74 chr12 113492311 113497546 DTX1 TSS
#> 75 chr12 122456912 122464036 BCL7A TSS
#> 76 chr13 46957278 46963342 C13orf18 TSS
#> 77 chr14 69257848 69259739 ZFP36L1 TSS
#> 78 chr14 94940587 94942549 SERPINA9 TSS
#> 79 chr14 96179535 96180366 TCL1A TSS
#> 80 chr15 59658991 59671152 MYO1E TSS
#> 81 chr16 10970795 10975465 CIITA TSS
#> 82 chr16 11347512 11350007 SOCS1 intron-1
#> 83 chr16 27322895 27329423 IL4R TSS
#> 84 chr16 85931918 85933977 IRF8 TSS
#> 85 chr17 3597616 3599572 P2RX5 TSS
#> 86 chr17 56407732 56410140 TSPOAP1 intergenic
#> 87 chr17 75424734 75440956 SEPT9 intronic-1
#> 88 chr17 75443766 75451177 SEPT9 intronic-2
#> 89 chr17 75453203 75471471 SEPT9 intronic-3
#> 90 chr17 79478289 79479959 ACTG1 intronic
#> 91 chr18 60796984 60814103 BCL2 intronic
#> 92 chr18 60982728 60988342 BCL2 TSS
#> 93 chr19 10340142 10341764 S1PR2 TSS
#> 94 chr19 16434978 16439011 KLF2 TSS
#> 95 chr19 19279635 19281441 MEF2B TSS
#> 96 chr20 46128611 46138099 NCOA3 TSS
#> 97 chr20 49120482 49140477 PTPN1 TSS
#> 98 chr21 26934372 26937651 MIR155HG TSS
#> 99 chr22 23229554 23232042 IGLL5 TSS
#> 100 chrX 12993308 12994511 TMSB4X intronic
#> 101 chrX 48774756 48776255 PIM2 TSS
#> regulatory_comment regions
#> 1 <NA> chr1:6661482-6662702
#> 2 <NA> chr1:23885584-23885835
#> 3 <NA> chr1:150550814-150552135
#> 4 active_promoter chr1:203274698-203275778
#> 5 weak_enhancer chr1:226864857-226873452
#> 6 active_promoter chr1:226920563-226927885
#> 7 enhancer chr1:226921088-226927982
#> 8 active_promoter chr2:60773789-60783486
#> 9 enhancer chr2:96808901-96811913
#> 10 active_promoter chr2:111874854-111882174
#> 11 weak_promoter chr2:136874728-136875461
#> 12 active_promoter-strong_enhancer chr3:16546433-16556786
#> 13 active_promoter chr3:32020518-32024930
#> 14 active_promoter chr3:71104986-71119518
#> 15 active_promoter chr3:71176427-71180960
#> 16 active_promoter chr3:71350633-71357665
#> 17 intronic chr3:71503103-71504870
#> 18 intronic chr3:71550753-71551478
#> 19 active_promoter chr3:71626341-71635648
#> 20 active_promoter chr3:101546669-101547704
#> 21 active_promoter chr3:101568239-101569274
#> 22 active_promoter-strong_enhancer chr3:183269360-183274139
#> 23 active_promoter chr3:186739628-186740875
#> 24 strong_enhancer chr3:186782522-186784438
#> 25 <NA> chr3:187458526-187464632
#> 26 <NA> chr3:187771678-187801626
#> 27 <NA> chr3:187801627-187813568
#> 28 <NA> chr3:187813569-187955254
#> 29 <NA> chr3:187955255-187969220
#> 30 <NA> chr3:188377178-188401951
#> 31 <NA> chr3:188401952-188461879
#> 32 <NA> chr3:188461880-188474683
#> 33 <NA> chr3:188474684-188491248
#> 34 active_promoter chr4:40193105-40204231
#> 35 active_promoter chr5:88131209-88174373
#> 36 active_promoter chr5:88174374-88182243
#> 37 active_promoter chr5:88182244-88206620
#> 38 active_promoter chr5:131823933-131826458
#> 39 active_promoter chr5:149790977-149792349
#> 40 active_promoter chr5:158500476-158532769
#> 41 active_promoter chr6:390572-394093
#> 42 active_promoter-strong_enhancer chr6:14118026-14120025
#> 43 enhancer chr6:31548325-31550717
#> 44 active_promoter chr6:37138104-37139804
#> 45 <NA> chr6:90981034-91016134
#> 46 active_promoter chr6:134487960-134499859
#> 47 <NA> chr6:159237903-159240216
#> 48 active_promoter chr7:5568297-5570856
#> 49 strong_enhancer chr8:11347723-11355318
#> 50 active_promoter chr8:128748352-128749427
#> 51 intronic chr9:37023396-37027663
#> 52 active_promoter chr9:37029849-37037154
#> 53 intronic chr9:37192080-37207549
#> 54 intronic chr9:37275952-37306152
#> 55 intronic chr9:37312655-37328260
#> 56 intronic chr9:37329706-37340398
#> 57 enhancer chr9:37369209-37372160
#> 58 enhancer chr9:37382267-37385854
#> 59 enhancer chr9:37395932-37409239
#> 60 active_promoter chr9:37423010-37425279
#> 61 active_promoter chr10:127578912-127591133
#> 62 <NA> chr11:9595246-9599502
#> 63 active_promoter chr11:35156769-35164248
#> 64 active_promoter chr11:60223385-60225310
#> 65 enhancer chr11:65190369-65192380
#> 66 enhancer chr11:65265237-65268359
#> 67 active_promoter chr11:102188170-102190077
#> 68 active_promoter chr11:111248078-111250224
#> 69 enhancer chr11:128339774-128345731
#> 70 active_promoter chr11:128388492-128394163
#> 71 strong_enhancer chr12:11796001-11812968
#> 72 active_promoter chr12:25204045-25213569
#> 73 active_promoter chr12:92537999-92539598
#> 74 <NA> chr12:113492311-113497546
#> 75 poised_promoter chr12:122456912-122464036
#> 76 active_promoter chr13:46957278-46963342
#> 77 active_promoter chr14:69257848-69259739
#> 78 <NA> chr14:94940587-94942549
#> 79 active_promoter chr14:96179535-96180366
#> 80 <NA> chr15:59658991-59671152
#> 81 active_promoter-strong_enhancer chr16:10970795-10975465
#> 82 enhancer chr16:11347512-11350007
#> 83 active_promoter chr16:27322895-27329423
#> 84 active_promoter chr16:85931918-85933977
#> 85 active_promoter chr17:3597616-3599572
#> 86 enhancer chr17:56407732-56410140
#> 87 active_promoter chr17:75424734-75440956
#> 88 active_promoter chr17:75443766-75451177
#> 89 active_promoter chr17:75453203-75471471
#> 90 <NA> chr17:79478289-79479959
#> 91 strong_enhancer chr18:60796984-60814103
#> 92 active_promoter chr18:60982728-60988342
#> 93 active_promoter chr19:10340142-10341764
#> 94 intronic chr19:16434978-16439011
#> 95 active_promoter chr19:19279635-19281441
#> 96 active_promoter chr20:46128611-46138099
#> 97 active_promoter chr20:49120482-49140477
#> 98 active_promoter chr21:26934372-26937651
#> 99 <NA> chr22:23229554-23232042
#> 100 active_promoter chrX:12993308-12994511
#> 101 active_promoter chrX:48774756-48776255
#> name
#> 1 KLHL21-TSS
#> 2 ID3-TSS
#> 3 MCL1-intronic
#> 4 BTG2-intronic
#> 5 ITPKB-intronic
#> 6 ITPKB-TSS
#> 7 ITPKB-intron-1
#> 8 BCL11A-TSS
#> 9 DUSP2-intron-1
#> 10 BCL2L11-TSS
#> 11 CXCR4-intronic
#> 12 RFTN1-TSS
#> 13 OSBPL10-TSS-1
#> 14 FOXP1-TSS-4
#> 15 FOXP1-TSS-3
#> 16 FOXP1-TSS-2
#> 17 FOXP1-intron-1b
#> 18 FOXP1-intron-1a
#> 19 FOXP1-TSS-1
#> 20 NFKBIZ-TSS-1
#> 21 NFKBIZ-TSS-2
#> 22 KLHL6-TSS
#> 23 ST6GAL1-TSS-1
#> 24 ST6GAL1-intronic-1
#> 25 BCL6-TSS
#> 26 LPP-TSS-1
#> 27 LPP-TSS-2
#> 28 LPP-TSS-3
#> 29 LPP-TSS-4
#> 30 LPP-intronic-1
#> 31 LPP-intronic-2
#> 32 LPP-intronic-3
#> 33 LPP-intronic-4
#> 34 RHOH-TSS
#> 35 MEF2C-TSS-3
#> 36 MEF2C-TSS-2
#> 37 MEF2C-TSS-1
#> 38 IRF1-TSS
#> 39 CD74-TSS
#> 40 EBF1-TSS
#> 41 IRF4-TSS
#> 42 CD83-TSS
#> 43 LTB-intron-1
#> 44 PIM1-TSS
#> 45 BACH2-TSS
#> 46 SGK1-TSS-1
#> 47 EZR-TSS
#> 48 ACTB-TSS
#> 49 BLK-TSS
#> 50 MYC-TSS
#> 51 PAX5-intron-1
#> 52 PAX5-TSS-1
#> 53 ZCCHC7-intron-4
#> 54 ZCCHC7-intron-2
#> 55 ZCCHC7-intron-3
#> 56 ZCCHC7-intron-1
#> 57 PAX5-distal-enhancer-1
#> 58 PAX5-distal-enhancer-2
#> 59 PAX5-distal-enhancer-3
#> 60 GRHPR-TSS
#> 61 FANK1-TSS
#> 62 WEE1-intronic
#> 63 CD44-TSS
#> 64 MS4A1-TSS
#> 65 NEAT1-ncRNA
#> 66 MALAT1-ncRNA
#> 67 BIRC3-TSS
#> 68 POU2AF1-TSS
#> 69 ETS1-introns
#> 70 ETS1-TSS-2
#> 71 ETV6-TSS
#> 72 LRMP-TSS
#> 73 BTG1-TSS
#> 74 DTX1-TSS
#> 75 BCL7A-TSS
#> 76 C13orf18-TSS
#> 77 ZFP36L1-TSS
#> 78 SERPINA9-TSS
#> 79 TCL1A-TSS
#> 80 MYO1E-TSS
#> 81 CIITA-TSS
#> 82 SOCS1-intron-1
#> 83 IL4R-TSS
#> 84 IRF8-TSS
#> 85 P2RX5-TSS
#> 86 TSPOAP1-intergenic
#> 87 SEPT9-intronic-1
#> 88 SEPT9-intronic-2
#> 89 SEPT9-intronic-3
#> 90 ACTG1-intronic
#> 91 BCL2-intronic
#> 92 BCL2-TSS
#> 93 S1PR2-TSS
#> 94 KLF2-TSS
#> 95 MEF2B-TSS
#> 96 NCOA3-TSS
#> 97 PTPN1-TSS
#> 98 MIR155HG-TSS
#> 99 IGLL5-TSS
#> 100 TMSB4X-intronic
#> 101 PIM2-TSS
#> Joining with `by = join_by(sample_id)`
#> Joining with `by = join_by(start)`
#> Joining with `by = join_by(sample_id)`