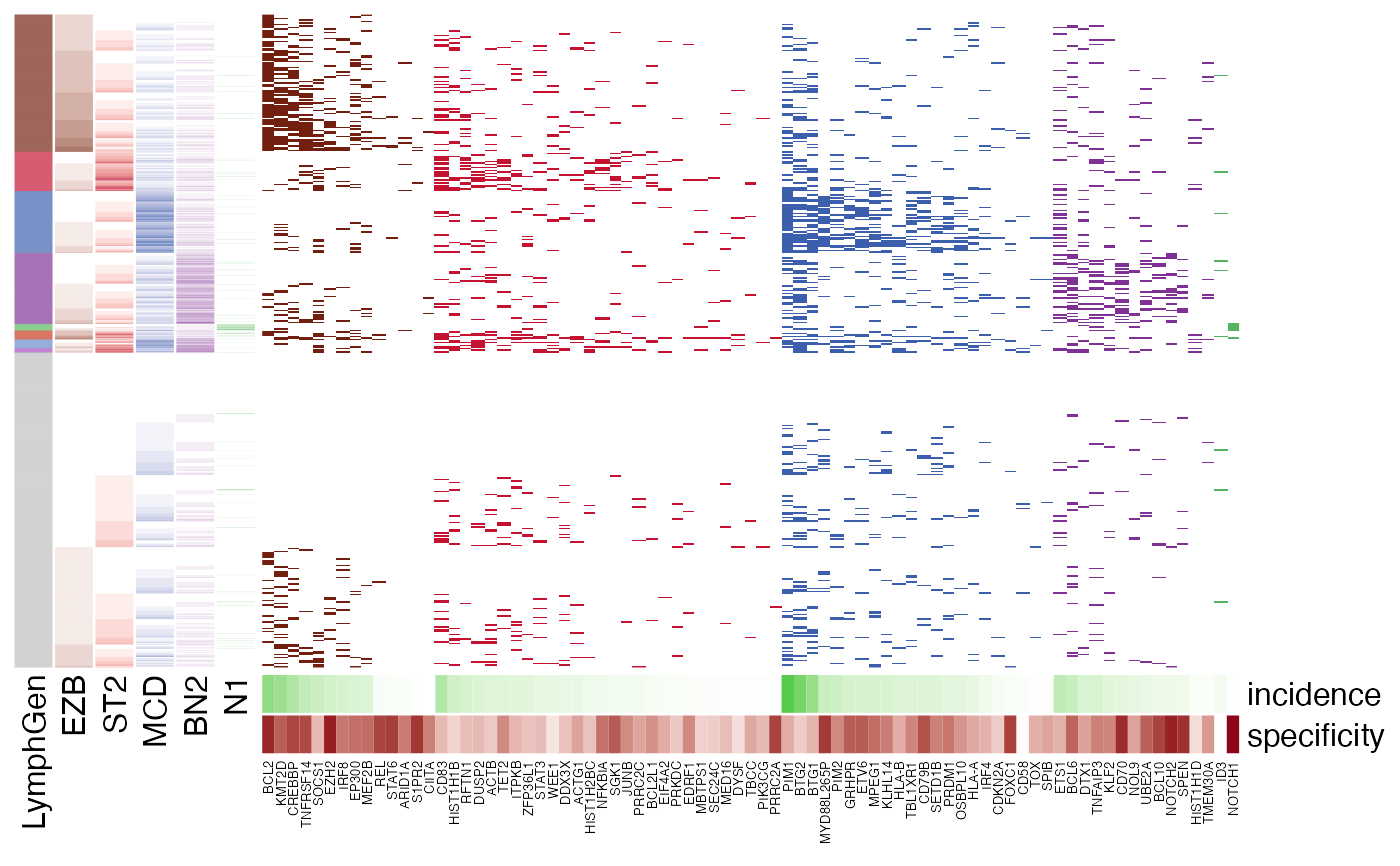
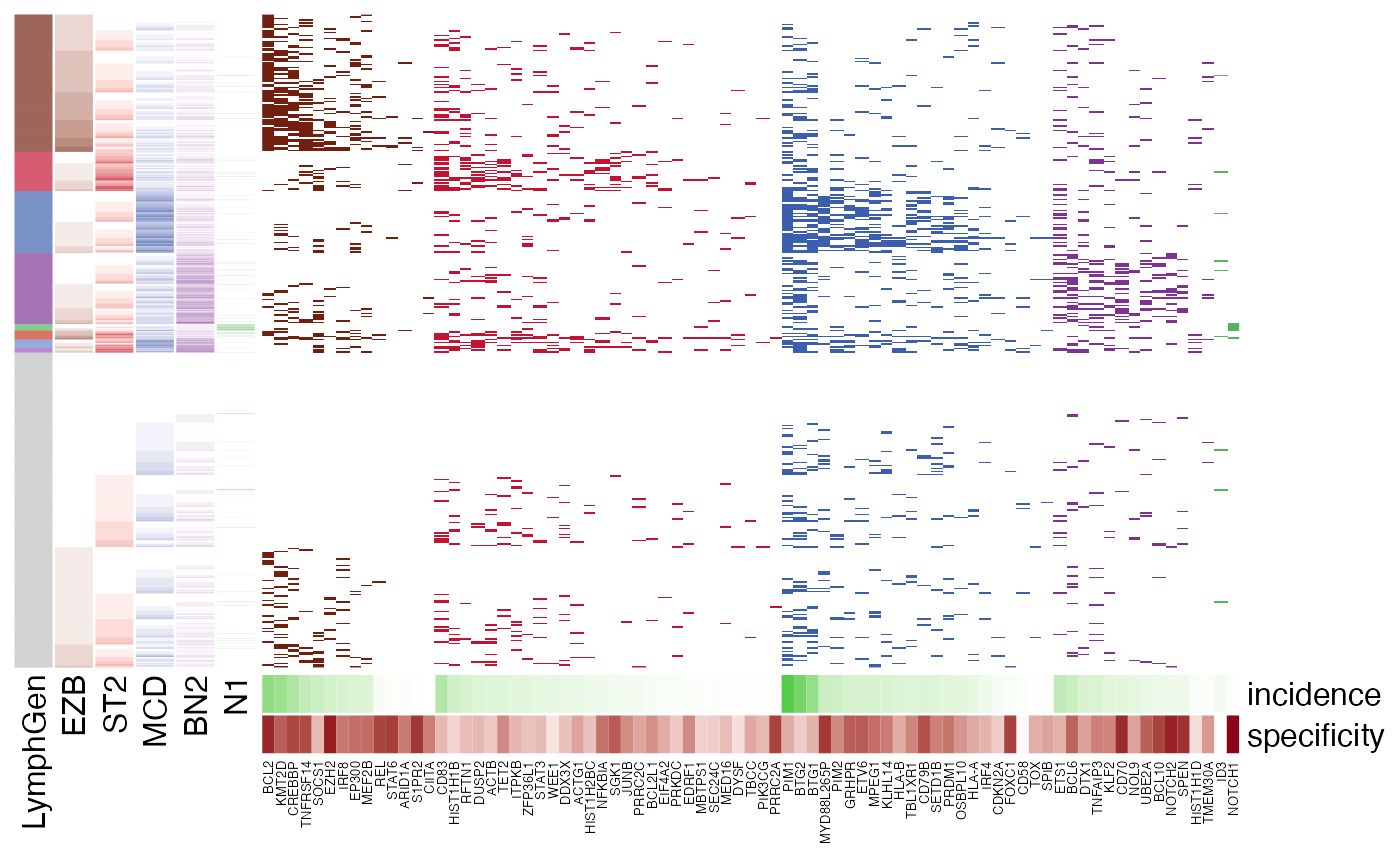
LymphGen feature visualization.
pretty_lymphoplot.RdMake a heatmap showing the frequency of LymphGen features across a cohort of samples.
Usage
pretty_lymphoplot(
these_samples_metadata,
flavour = "with_cnvs.with_sv.with_A53",
lymphgen_all,
with_A53 = FALSE,
show_side_annotation = FALSE,
verbose = FALSE
)Arguments
- these_samples_metadata
A data frame containing the metadata for your samples (LymphGen result will be subset to the sample_id in this data frame).
- flavour
Required if the output of get_lymphgen has not been provided.
- lymphgen_all
The list returned by get_lymphgen.
- with_A53
Set to FALSE if you are using LymphGen results without A53. Recommended option.
- show_side_annotation
Set to TRUE if you want the default ComplexHeatmap annotations on the sides of your heatmap. Default is FALSE.
- verbose
Set to TRUE to print helpful messages. Useful for debugging. Default is FALSE (not verbose).
Examples
meta_df = suppressMessages(get_gambl_metadata()) %>%
dplyr::filter(seq_type %in% c("genome", "capture")) %>%
dplyr::filter(pathology == "DLBCL")
pretty_lymphoplot(meta_df, show_side_annotation = TRUE)
#> Joining with `by = join_by(Sample.Name, BTG2)`
#> Joining with `by = join_by(Sample.Name)`
#> Joining with `by = join_by(Sample.Name, SOCS1)`
#> Joining with `by = join_by(Sample.Name)`
#> Joining with `by = join_by(sample_id)`
#> Warning: The input is a data frame-like object, convert it to a matrix.
#> Warning: Note: not all columns in the data frame are numeric. The data frame
#> will be converted into a character matrix.
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> 'magick' package is suggested to install to give better rasterization.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
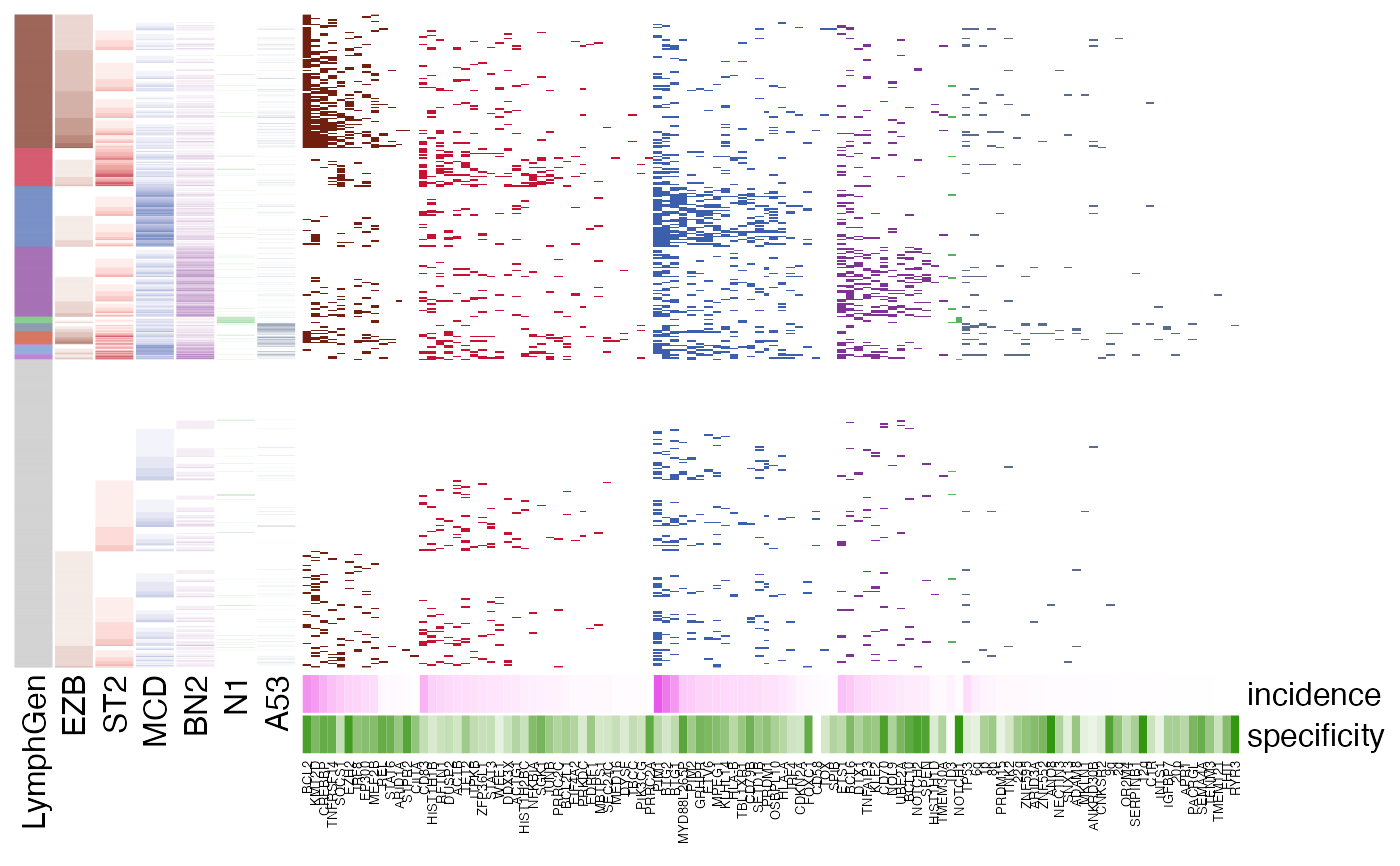
 pretty_lymphoplot(
meta_df,
show_side_annotation = TRUE,
flavour = "with_cnvs.with_sv.no_A53"
)
#> NO A53
#> Joining with `by = join_by(Sample.Name)`
#> Joining with `by = join_by(Sample.Name, BTG2)`
#> Joining with `by = join_by(Sample.Name)`
#> Joining with `by = join_by(Sample.Name, SOCS1)`
#> Running in the mode without A53 ...
#> Joining with `by = join_by(sample_id)`
#> Warning: The input is a data frame-like object, convert it to a matrix.
#> Warning: Note: not all columns in the data frame are numeric. The data frame
#> will be converted into a character matrix.
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> 'magick' package is suggested to install to give better rasterization.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
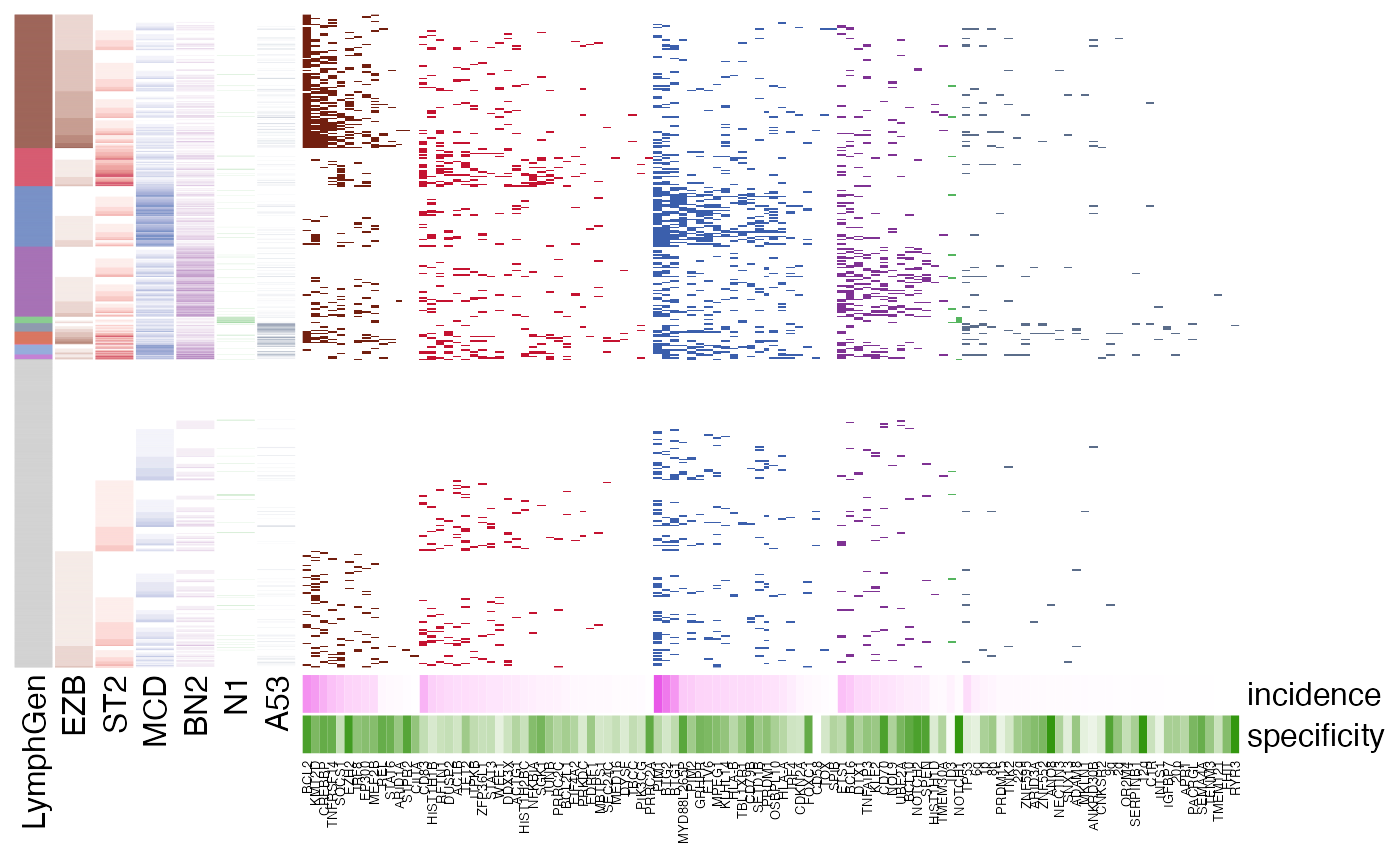
pretty_lymphoplot(
meta_df,
show_side_annotation = TRUE,
flavour = "with_cnvs.with_sv.no_A53"
)
#> NO A53
#> Joining with `by = join_by(Sample.Name)`
#> Joining with `by = join_by(Sample.Name, BTG2)`
#> Joining with `by = join_by(Sample.Name)`
#> Joining with `by = join_by(Sample.Name, SOCS1)`
#> Running in the mode without A53 ...
#> Joining with `by = join_by(sample_id)`
#> Warning: The input is a data frame-like object, convert it to a matrix.
#> Warning: Note: not all columns in the data frame are numeric. The data frame
#> will be converted into a character matrix.
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> 'magick' package is suggested to install to give better rasterization.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.